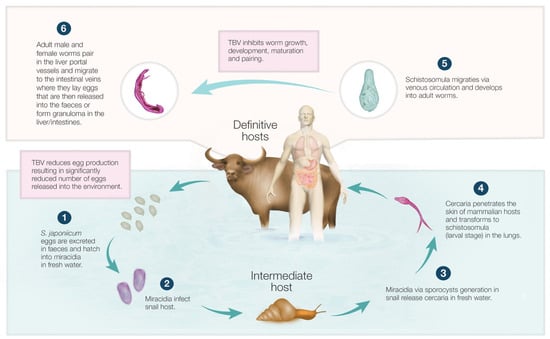
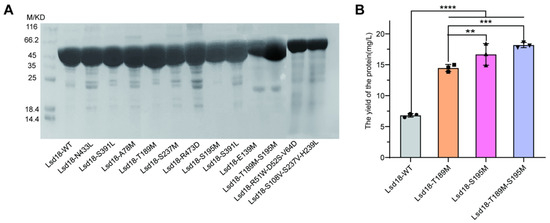
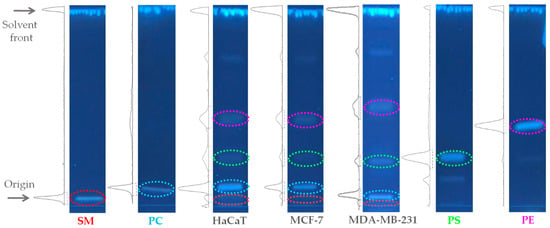
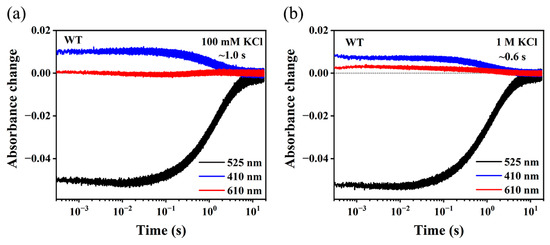
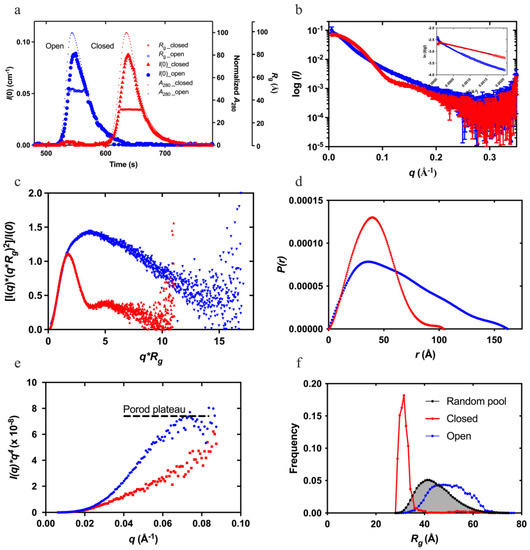
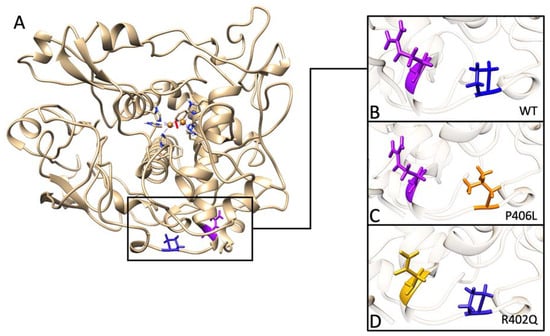
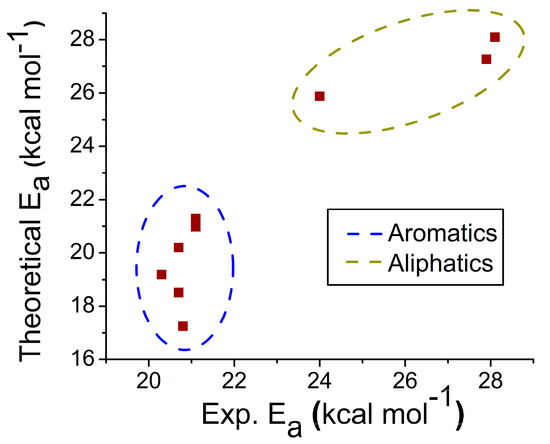
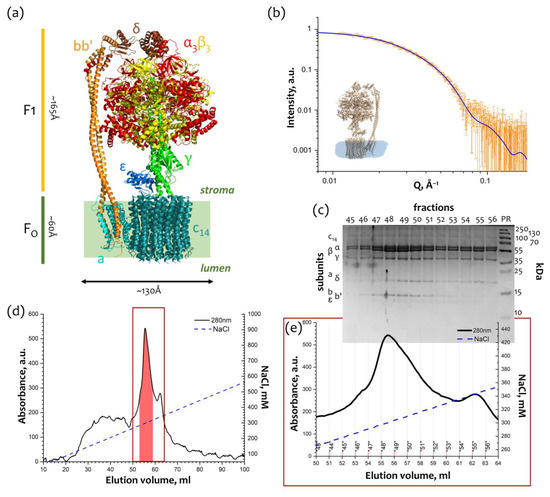
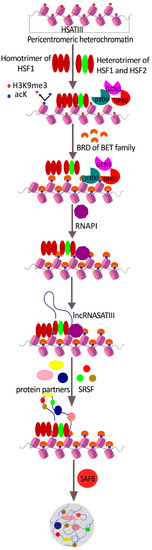
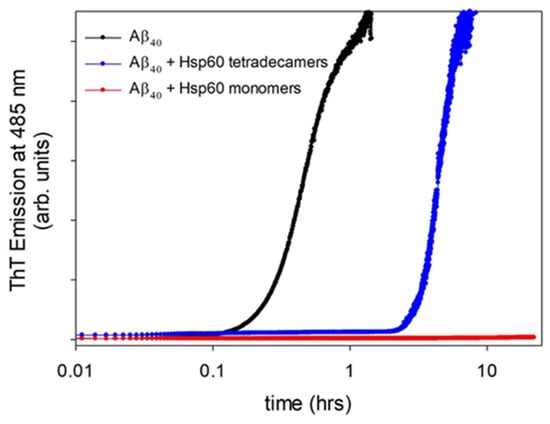
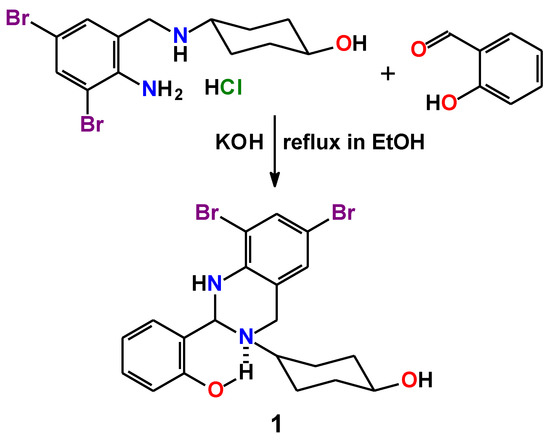
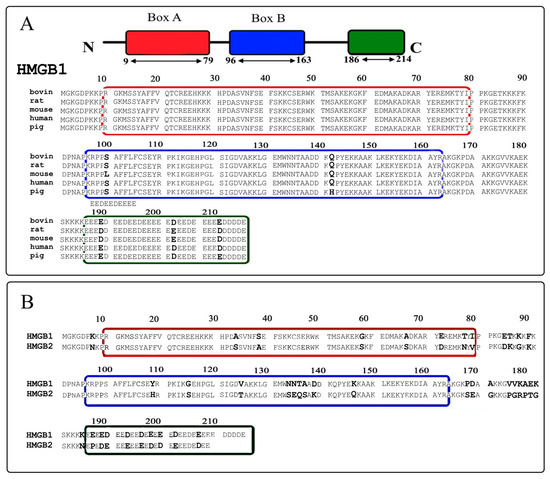
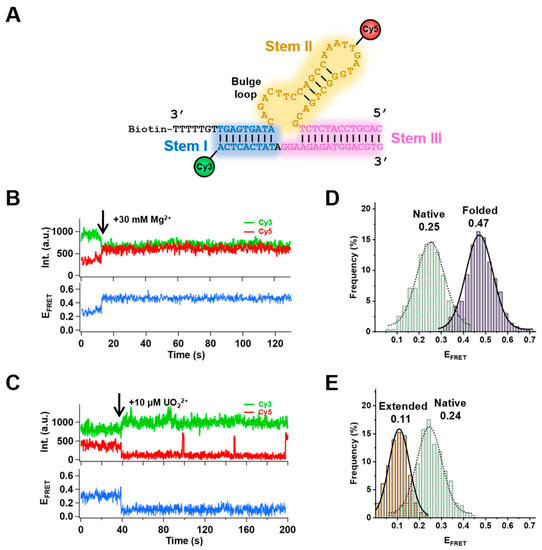
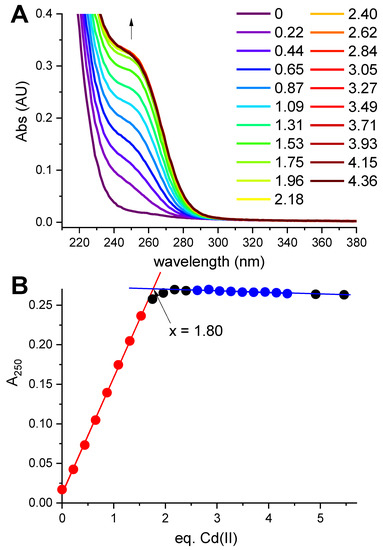
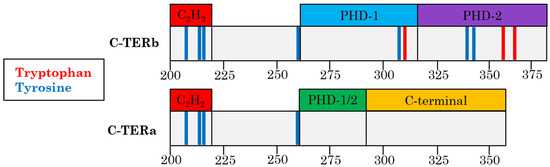
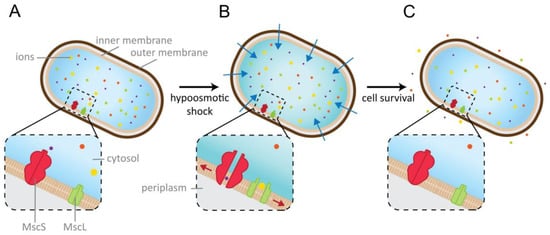
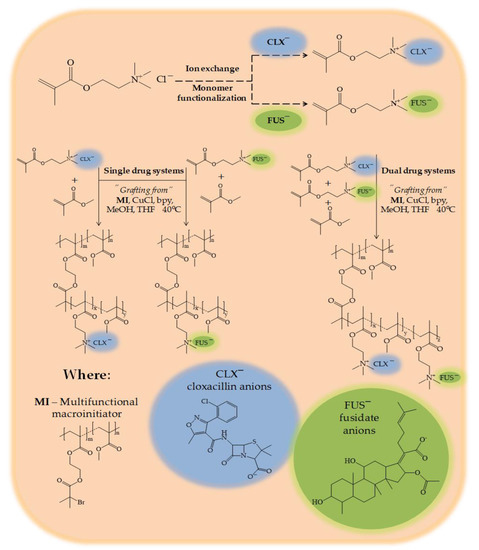
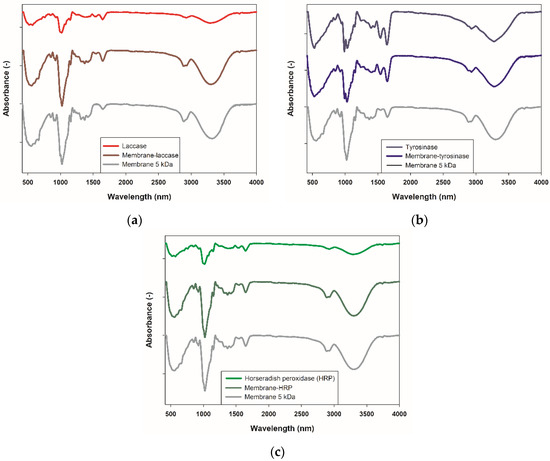
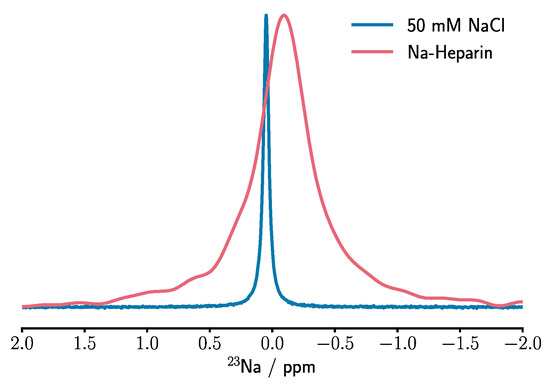
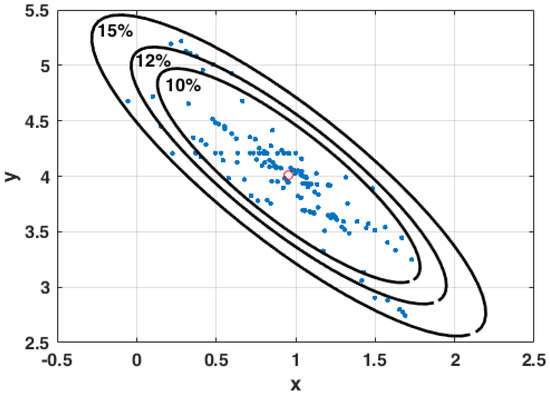
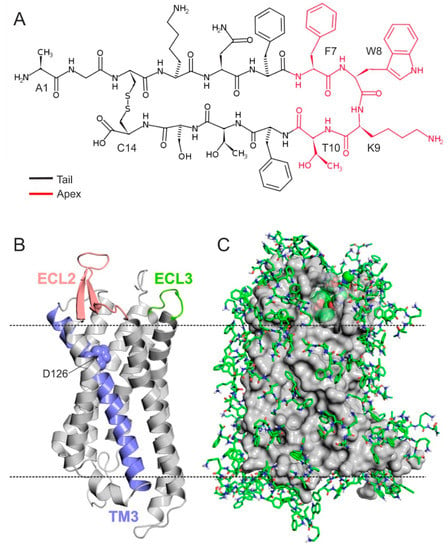
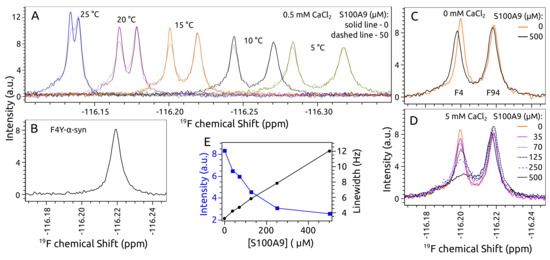
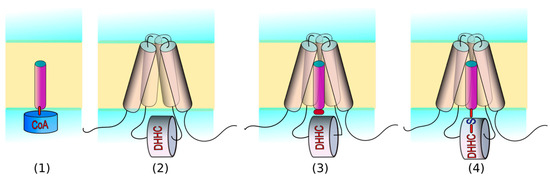
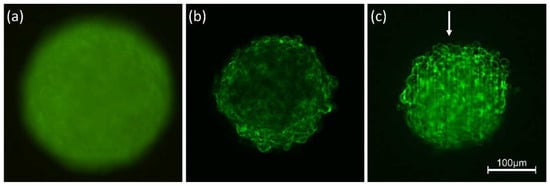
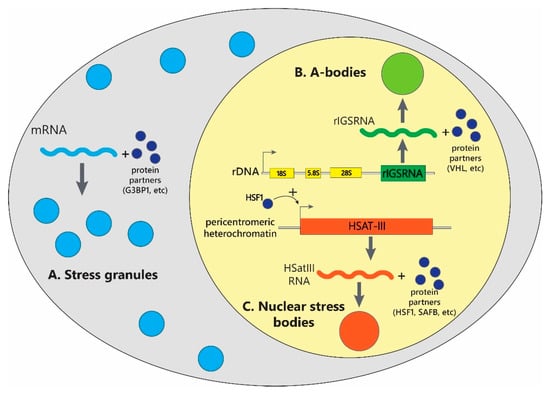
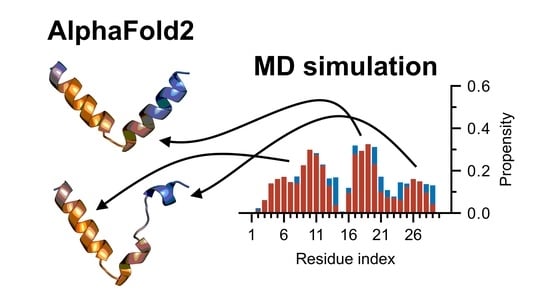
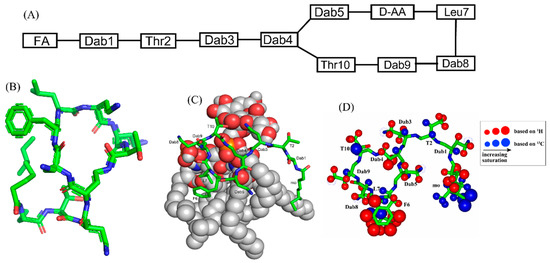
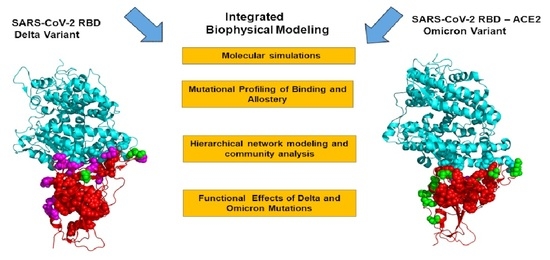
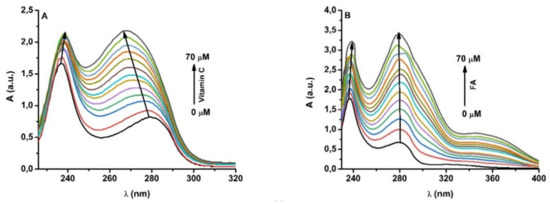
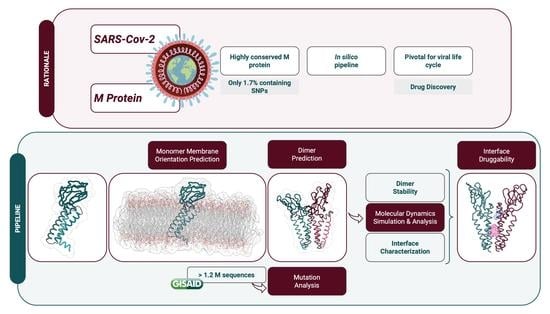
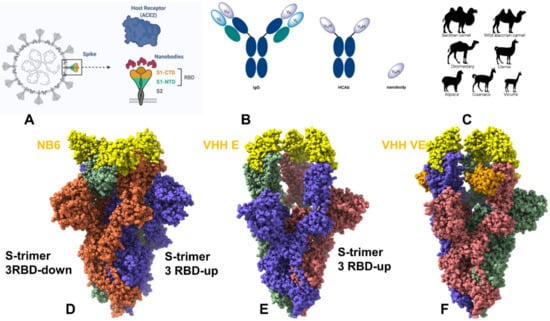
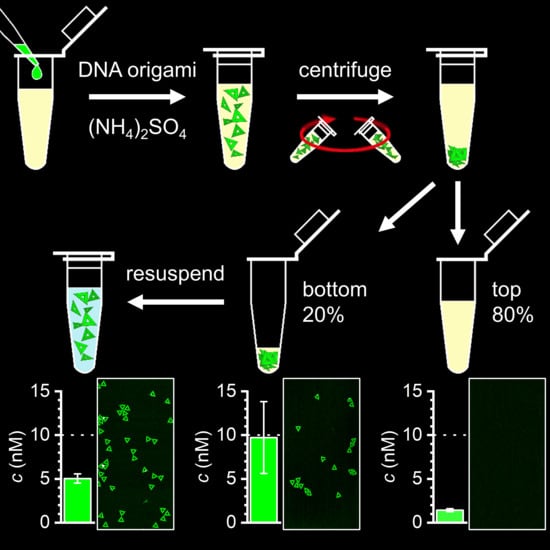
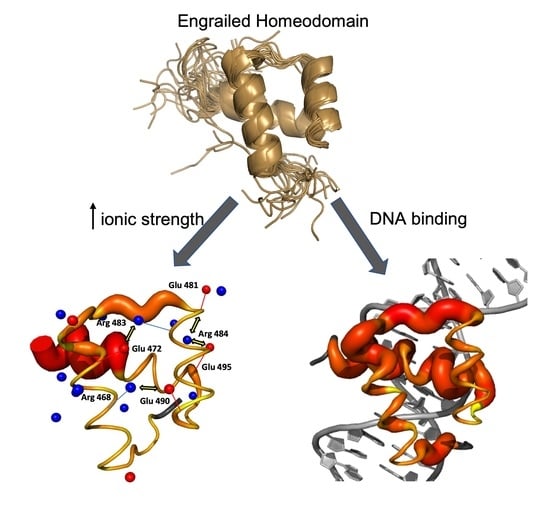
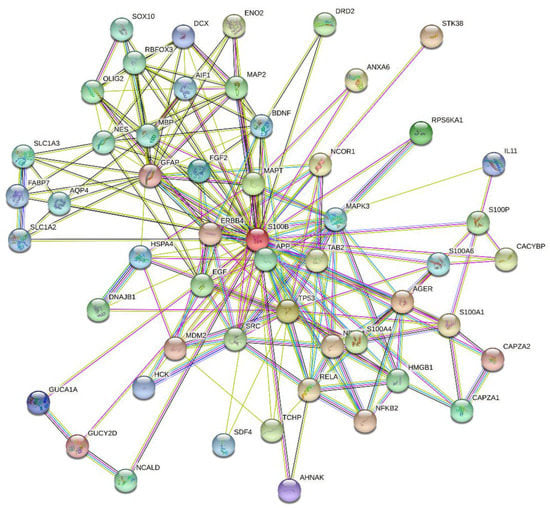
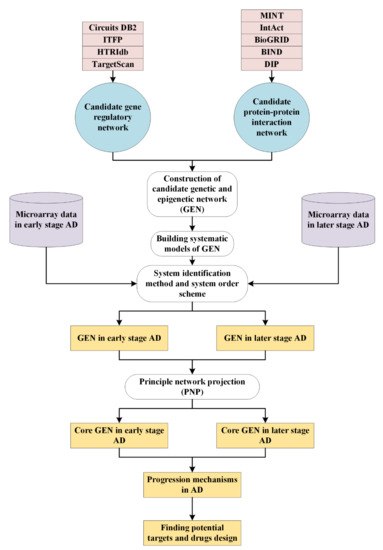
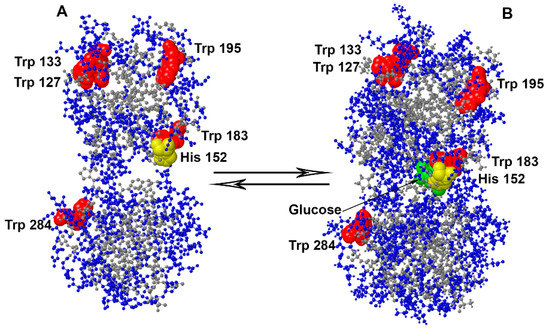
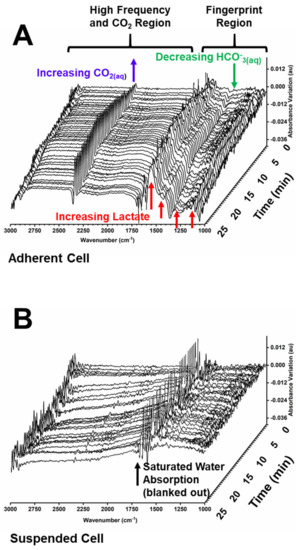
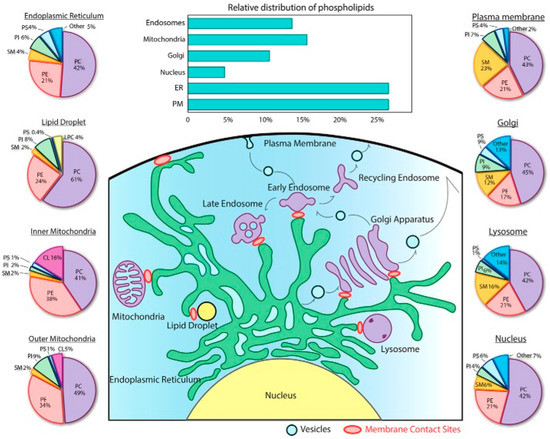
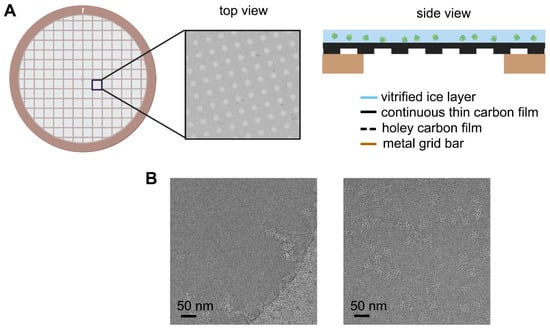
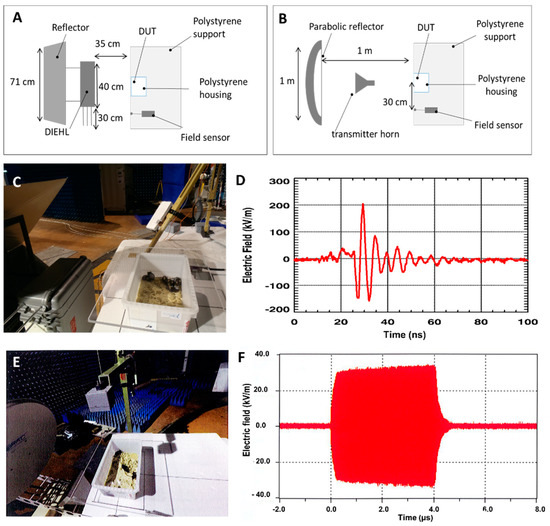
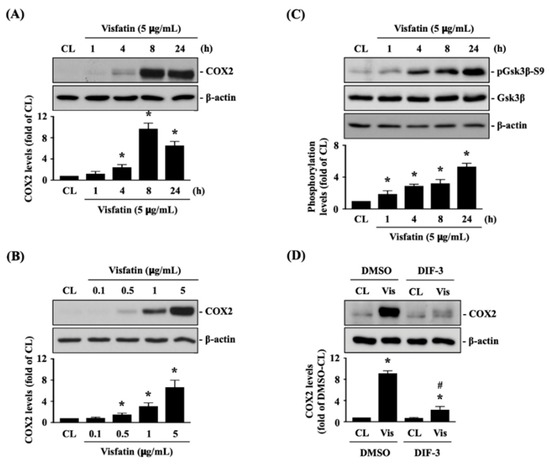
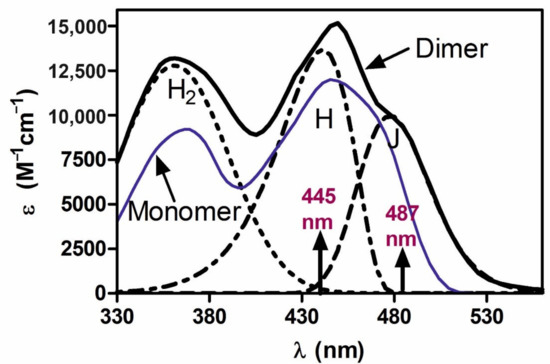
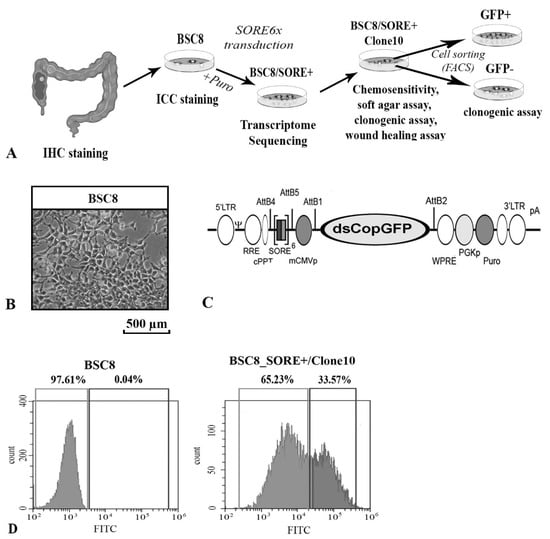
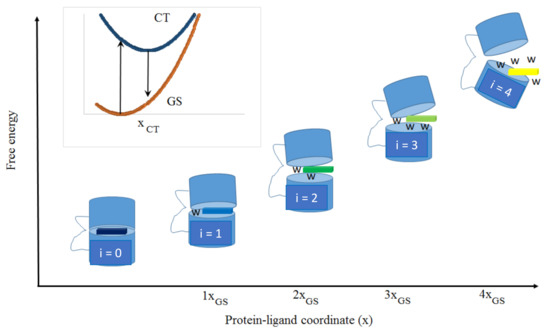
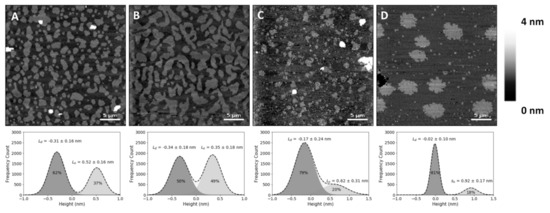
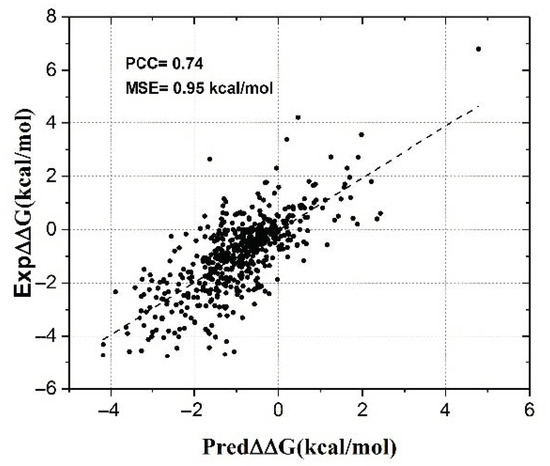
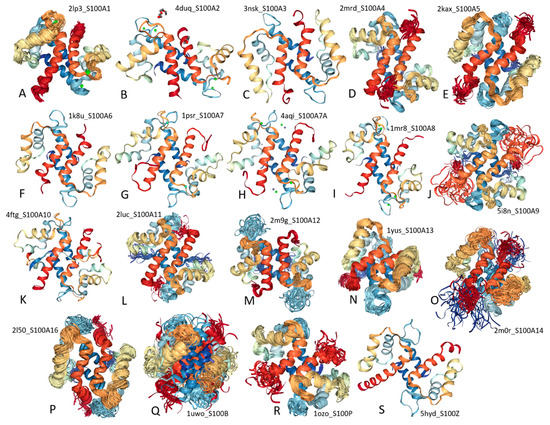
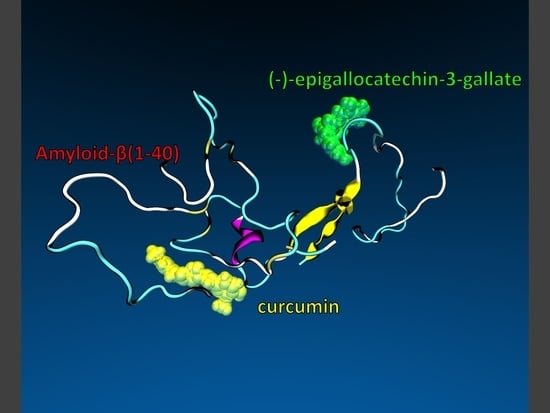
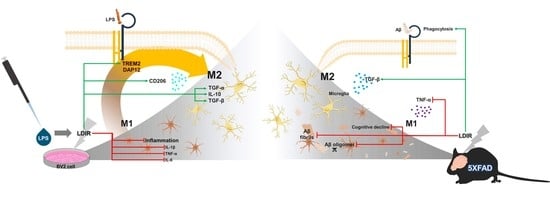
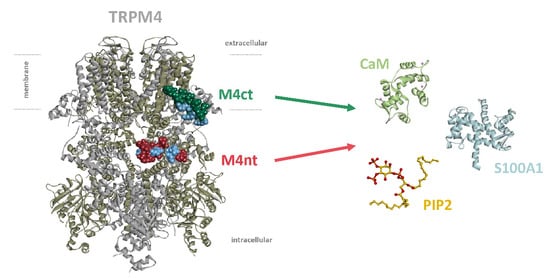
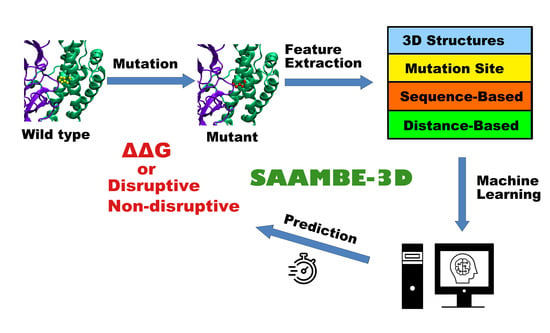
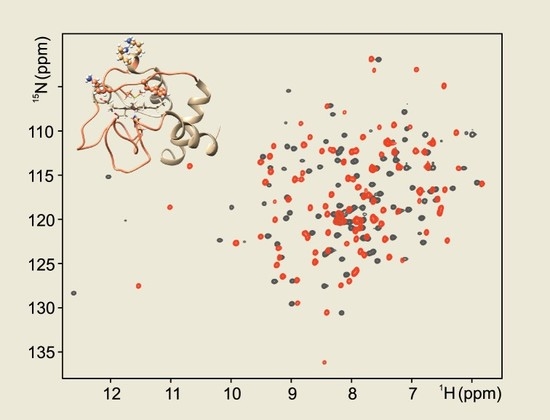
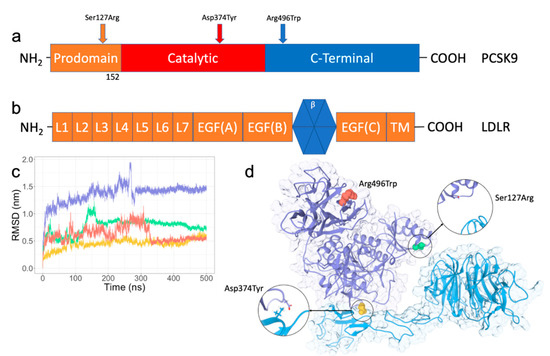
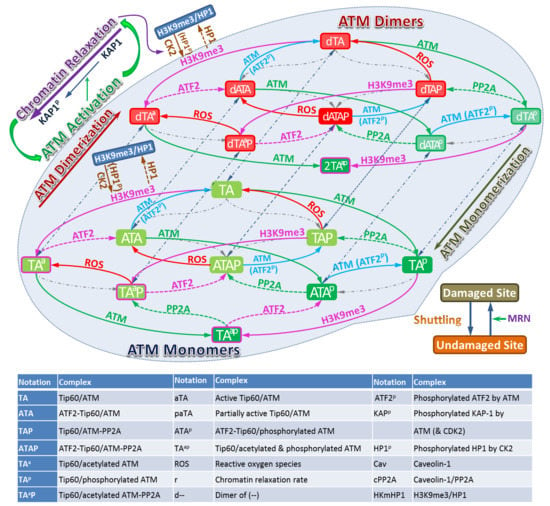
Feature Papers in Molecular Biophysics
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Biophysics".
Viewed by 667364Editors
Interests: biomimetic systems; complex mixture modelling and spectroscopic studies of biological; synthetic and hybrid systems
Special Issues, Collections and Topics in MDPI journals
Interests: intrinsically disordered proteins; protein folding; protein misfolding; partially folded proteins; protein aggregation; protein structure; protein function; protein stability; protein biophysics; protein bioinformatics; conformational diseases; protein–ligand interactions; protein–protein interactions; liquid-liquid phase transitions
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
As follows from the title, this Topical Collection “Feature Papers in Molecular Biophysics” aims to collect high quality research articles, short communications, and review articles in all the fields of molecular biophysics. Since the aim of this Topical Collection is to illustrate, through selected works, frontier research in molecular biophysics, we encourage Editorial Board Members of the Molecular Biophysics Section of the International Journal of Molecular Sciences to contribute papers reflecting the latest progress in their research field, or to invite relevant experts and colleagues to do so.
Topics include, but are not limited to:
- molecular structure and dynamics
- nucleic acid structure and dynamics
- protein structure and dynamics
- membrane structure and dynamics
- biomimetic material structure and dynamics
- molecular simulations
- molecular modeling
- single molecule biophysics
- biophysical techniques in the study of biomacromolecular and biomimetic systems
- biomolecular interactions
- biomimetic material interactions
- macromolecular structure determination or prediction
- characterization of disordered proteins and their interactions
- computational biophysics
- bioinformatics
- biophysical and computational approaches to drug design and development
- advances in molecular biophysical methodologies as well as imaging techniques and data analysis
- application of biophysical methods
Prof. Dr. Ian A. Nicholls
Prof. Dr. Vladimir N. Uversky
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- biophysics
- molecular imprinting
- molecular simulations
- molecular structure
- molecular dynamics
- molecular mechanics
- thermodynamics
- biomolecular interactions
- protein structure and folding
- intrinsically disordered proteins
- structure prediction
- nucleic acid–protein interactions
- protein–membrane interactions
- protein–DNA interactions
- posttranslational modifications
- drug–receptor interactions
- protein design
- protein engineering
- protein–ligand binding
- transmembrane proteins
- chaperones
- enzymology
- molecular recognition
- molecular modeling
- membrane dynamics
- macromolecular structure and dynamics
- DNA structure and dynamics
- RNA structure
- genome structure
- structure–function relationships
- ion channels
- spectroscopic techniques
- biomolecular NMR
- inter-molecular interactions
- X-ray crystallography
- macromolecular crystallography
- crystal thermodynamics
- microcalorimetry
- transient kinetic techniques
- fluorescence imaging
- single-molecule microscopy
- statistical mechanics
- computer simulations
- computational modeling
- molecular modeling