Zooming into the Dark Side of Human Annexin-S100 Complexes: Dynamic Alliance of Flexible Partners
Abstract
1. Introduction
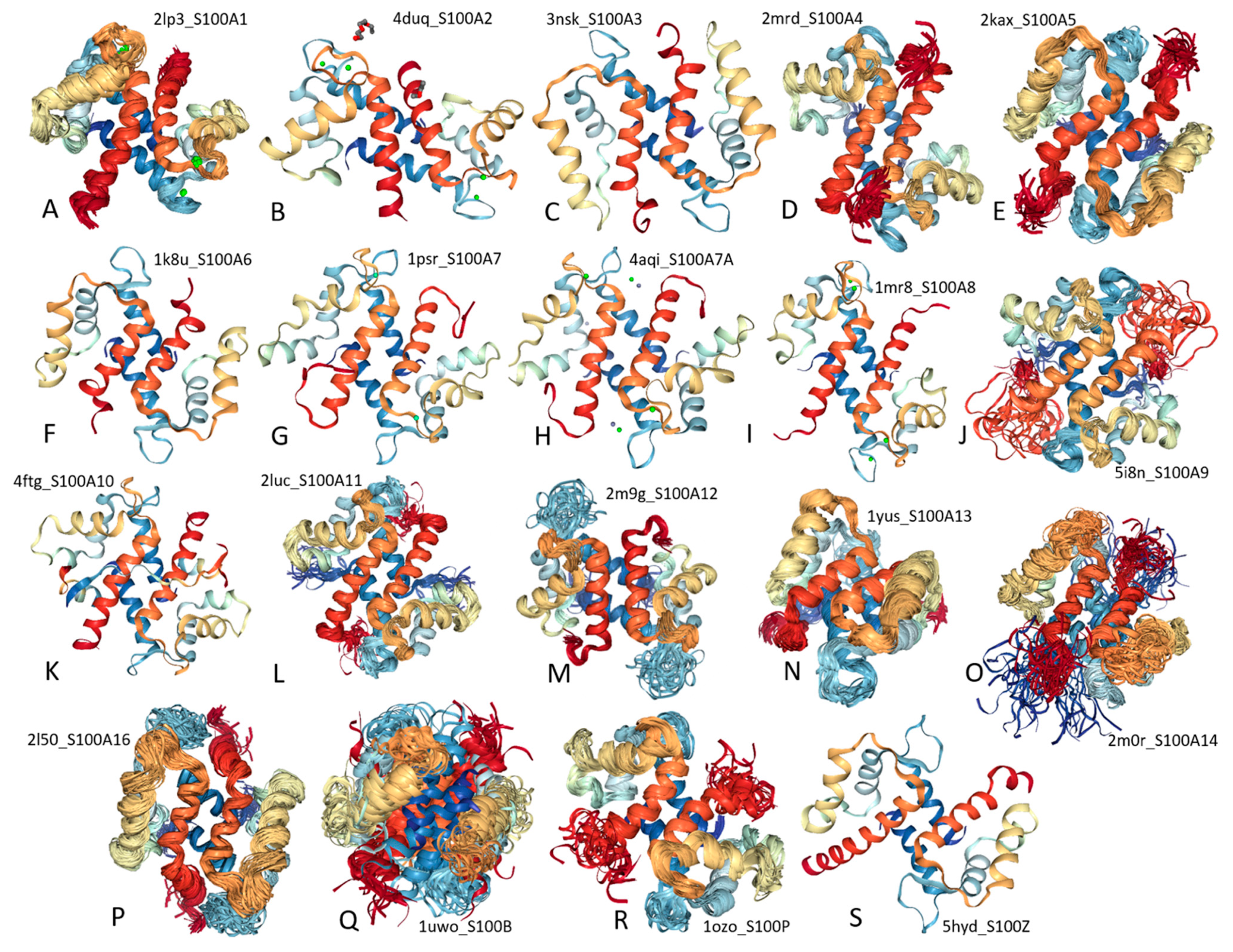
2. Structure and Functions of Human S100 Proteins
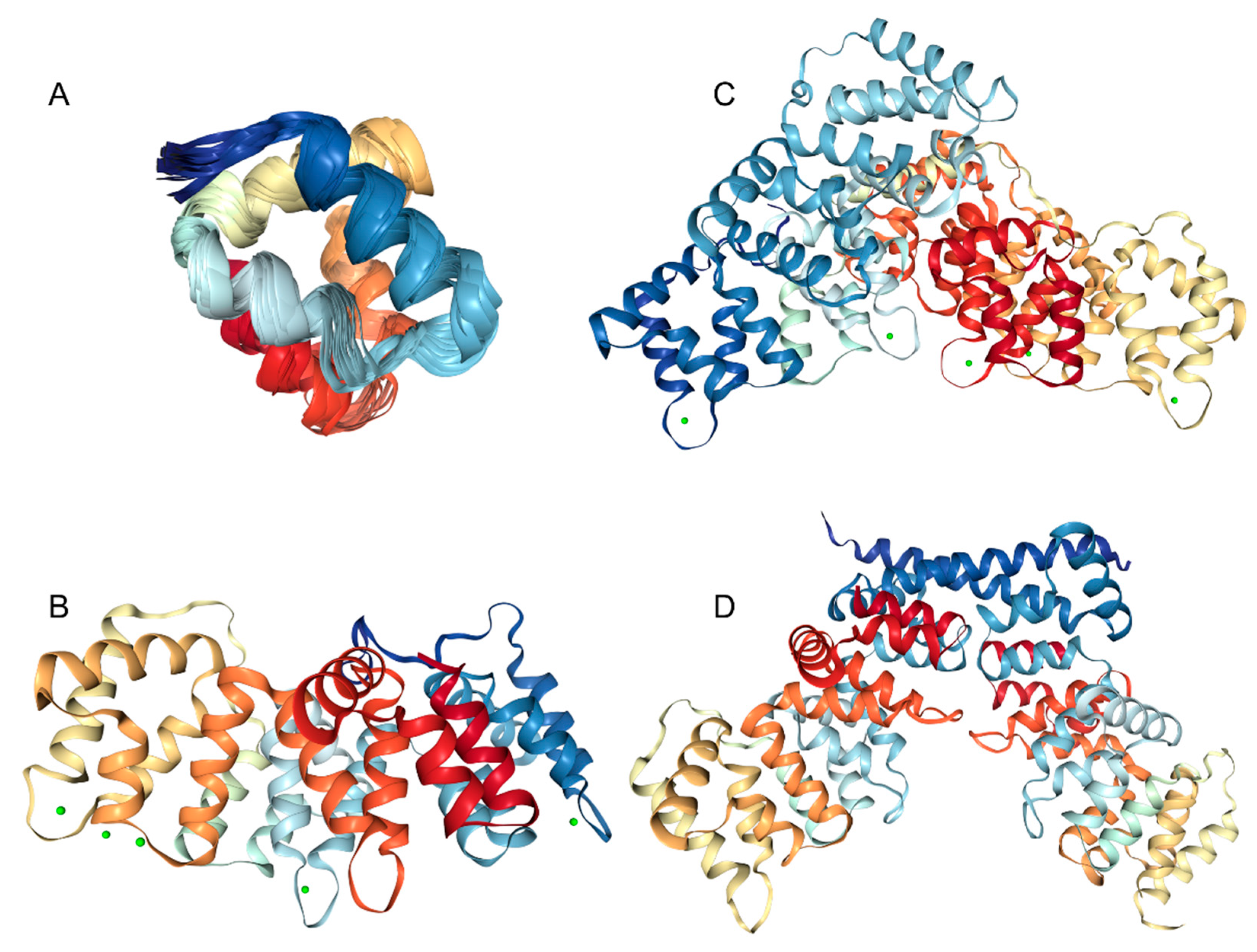
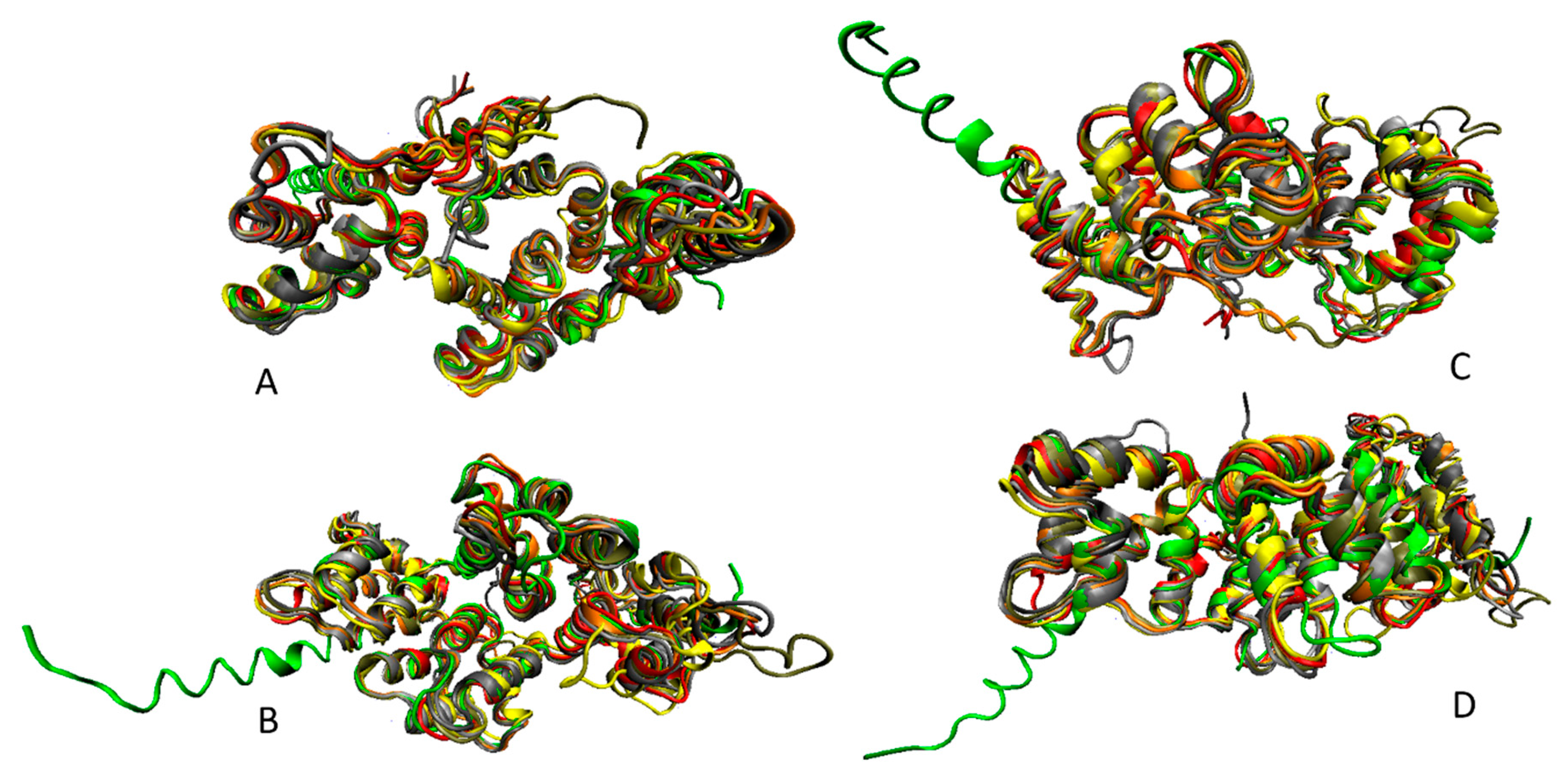
3. Structure and Functions of Human Annexins
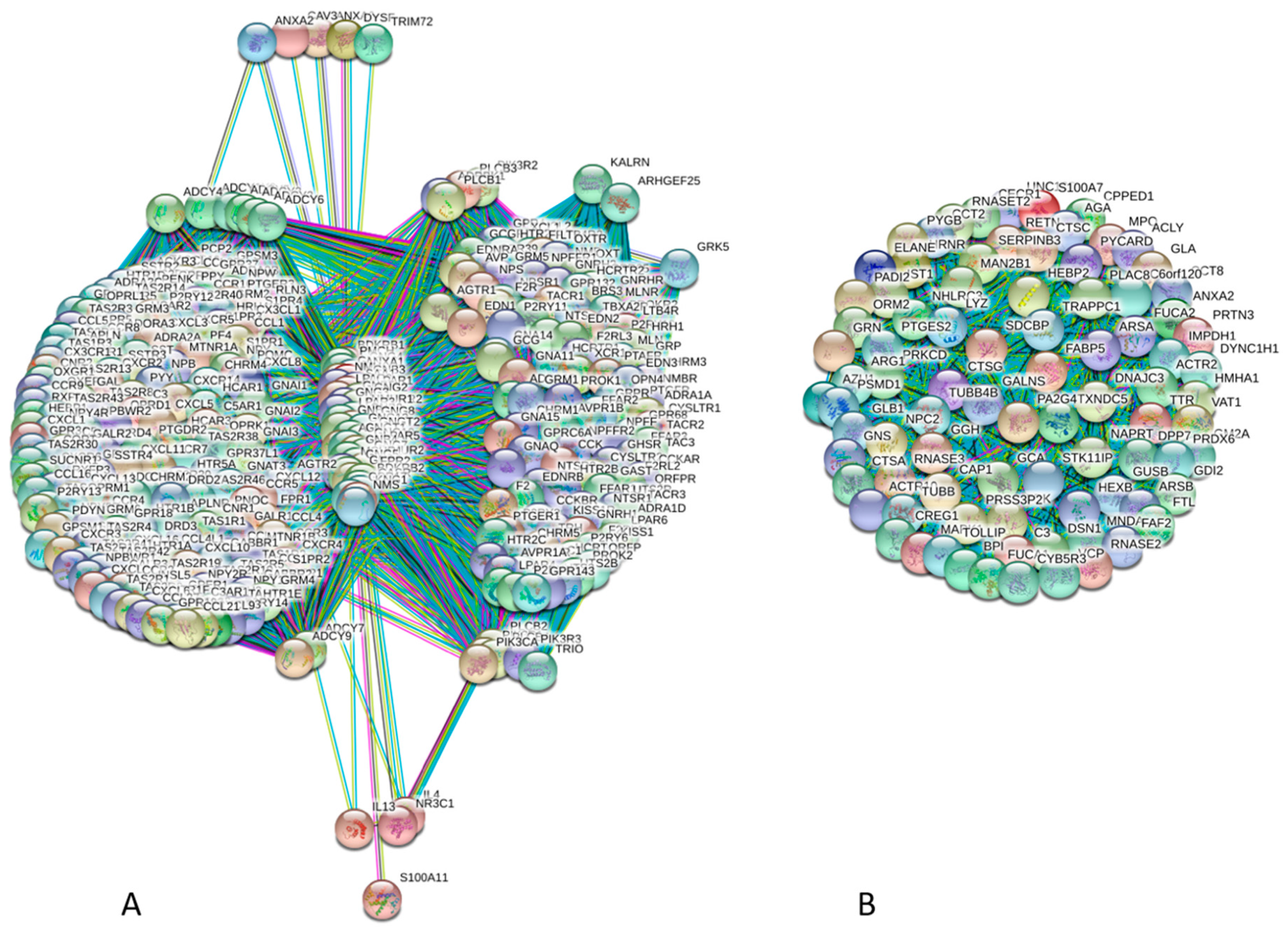
4. Interactability of Human Annexins and S100 Proteins
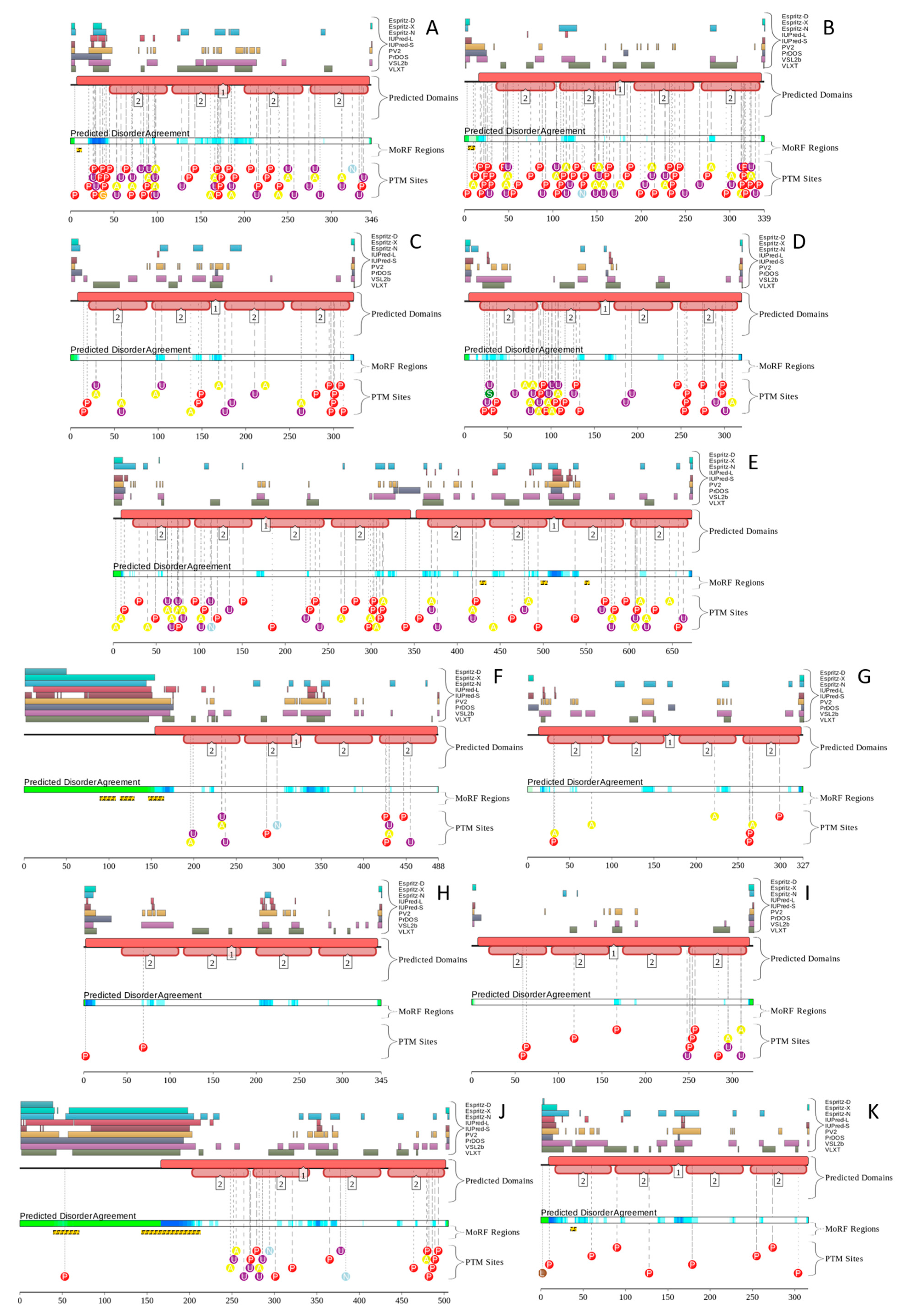
5. Intrinsic Disorder as a Common Denominator for Understanding the Multifunctionality of Annexins and S100 Proteins
6. Intrinsic Disorder and S100-Annexin Aomplexes
7. Conclusions: Structure-Function Continuum of Annexins and S100 Proteins; Proteoforms Originating from Intrinsic Disorder and Structural Polymorphism as a Clue for Understanding the Multifunctionality and Binding Promiscuity
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Miwa, N.; Uebi, T.; Kawamura, S. S100-annexin complexes—biology of conditional association. FEBS J. 2008, 275, 4945–4955. [Google Scholar] [CrossRef] [PubMed]
- Nockolds, C.E.; Kretsinger, R.H.; Coffee, C.J.; Bradshaw, R.A. Structure of a calcium-binding carp myogen. Proc. Natl. Acad. Sci. USA 1972, 69, 581–584. [Google Scholar] [CrossRef] [PubMed]
- Raynal, P.; Pollard, H.B. Annexins: The problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim. Biophys. Acta 1994, 1197, 63–93. [Google Scholar] [CrossRef]
- Gerke, V.; Moss, S.E. Annexins: From structure to function. Physiol. Rev. 2002, 82, 331–371. [Google Scholar] [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim. Biophys. Acta 2010, 1804, 1231–1264. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically disordered proteins: A 10-year recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013, 22, 693–724. [Google Scholar] [CrossRef] [PubMed]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef]
- Uversky, V.N. Dancing protein clouds: The strange biology and chaotic physics of intrinsically disordered proteins. J. Biol. Chem. 2016, 291, 6681–6688. [Google Scholar] [CrossRef]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef]
- Dunker, A.K.; Brown, C.J.; Obradovic, Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002, 62, 25–49. [Google Scholar]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell. Mol. Life Sci. 2003, 60, 1852–1871. [Google Scholar] [CrossRef]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta 2013, 1834, 932–951. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic. Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014, 23, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA 2006, 103, 8390–8395. [Google Scholar] [CrossRef]
- Buljan, M.; Chalancon, G.; Dunker, A.K.; Bateman, A.; Balaji, S.; Fuxreiter, M.; Babu, M.M. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 443–450. [Google Scholar] [CrossRef]
- Buljan, M.; Chalancon, G.; Eustermann, S.; Wagner, G.P.; Fuxreiter, M.; Bateman, A.; Babu, M.M. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol. Cell. 2012, 46, 871–883. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008, 37, 215–246. [Google Scholar] [CrossRef]
- Uversky, V.N.; Dave, V.; Iakoucheva, L.M.; Malaney, P.; Metallo, S.J.; Pathak, R.R.; Joerger, A.C. Pathological unfoldomics of uncontrolled chaos: Intrinsically disordered proteins and human diseases. Chem. Rev. 2014, 114, 6844–6879. [Google Scholar] [CrossRef]
- Uversky, V.N. Wrecked regulation of intrinsically disordered proteins in diseases: Pathogenicity of deregulated regulators. Front. Mol. Biosci. 2014, 1, 6. [Google Scholar] [CrossRef]
- Engelkamp, D.; Schafer, B.W.; Mattei, M.G.; Erne, P.; Heizmann, C.W. Six S100 genes are clustered on human chromosome 1q21: Identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc. Natl. Acad. Sci. USA 1993, 90, 6547–6551. [Google Scholar] [CrossRef] [PubMed]
- Schafer, B.W.; Wicki, R.; Engelkamp, D.; Mattei, M.G.; Heizmann, C.W. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: Rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics 1995, 25, 638–643. [Google Scholar] [CrossRef]
- Marenholz, I.; Lovering, R.C.; Heizmann, C.W. An update of the S100 nomenclature. Biochim. Biophys. Acta 2006, 1763, 1282–1283. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Sorci, G.; Riuzzi, F.; Arcuri, C.; Bianchi, R.; Brozzi, F.; Tubaro, C.; Giambanco, I. S100B’s double life: Intracellular regulator and extracellular signal. Biochim. Biophys. Acta 2009, 1793, 1008–1022. [Google Scholar] [CrossRef]
- Heizmann, C.W.; Fritz, G.; Schafer, B.W. S100 proteins: Structure, functions and pathology. Front. Biosci. 2002, 7, d1356–d1368. [Google Scholar] [PubMed]
- Heizmann, C.W.; Cox, J.A. New perspectives on S100 proteins: A multi-functional Ca(2+)-, Zn(2+)- and Cu(2+)-binding protein family. Biometals 1998, 11, 383–397. [Google Scholar] [CrossRef]
- Donato, R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell. Biol. 2001, 33, 637–668. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef]
- Santamaria-Kisiel, L.; Rintala-Dempsey, A.C.; Shaw, G.S. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 2006, 396, 201–214. [Google Scholar] [CrossRef]
- Schaub, M.C.; Heizmann, C.W. Calcium, troponin, calmodulin, S100 proteins: From myocardial basics to new therapeutic strategies. Biochem. Biophys. Res. Commun. 2008, 369, 247–264. [Google Scholar] [CrossRef]
- Permyakov, E. Metalloproteomics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; p. 786. [Google Scholar]
- Moroz, O.V.; Wilson, K.S.; Bronstein, I.B. The role of zinc in the S100 proteins: Insights from the X-ray structures. Amino Acids 2010, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Calderone, V.; Fragai, M.; Luchinat, C. Reviewing the crystal structure of S100Z and other members of the S100 family: Implications in calcium-regulated quaternary structure. Methods Mol. Biol. 2019, 1929, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Winningham-Major, F.; Staecker, J.L.; Barger, S.W.; Coats, S.; Van Eldik, L.J. Neurite extension and neuronal survival activities of recombinant S100 beta proteins that differ in the content and position of cysteine residues. J. Cell. Biol. 1989, 109, 3063–3071. [Google Scholar] [CrossRef]
- Heizmann, C.W.; Ackermann, G.E.; Galichet, A. Pathologies involving the S100 proteins and RAGE. Subcell. Biochem. 2007, 45, 93–138. [Google Scholar]
- Edgeworth, J.; Freemont, P.; Hogg, N. Ionomycin-regulated phosphorylation of the myeloid calcium-binding protein p14. Nature 1989, 342, 189–192. [Google Scholar] [CrossRef]
- Guignard, F.; Mauel, J.; Markert, M. Phosphorylation of myeloid-related proteins MRP-14 and MRP-8 during human neutrophil activation. Eur. J. Biochem. 1996, 241, 265–271. [Google Scholar] [CrossRef]
- Siegenthaler, G.; Roulin, K.; Chatellard-Gruaz, D.; Hotz, R.; Saurat, J.H.; Hellman, U.; Hagens, G. A heterocomplex formed by the calcium-binding proteins MRP8 (S100A8) and MRP14 (S100A9) binds unsaturated fatty acids with high affinity. J. Biol. Chem. 1997, 272, 9371–9377. [Google Scholar] [CrossRef]
- Kerkhoff, C.; Klempt, M.; Sorg, C. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9). Biochim. Biophys. Acta 1998, 1448, 200–211. [Google Scholar] [CrossRef]
- Heizmann, C.W. The multifunctional S100 protein family. Methods Mol. Biol. 2002, 172, 69–80. [Google Scholar] [CrossRef]
- Donato, R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim. Biophys. Acta 1999, 1450, 191–231. [Google Scholar] [CrossRef]
- Smith, S.P.; Shaw, G.S. A change-in-hand mechanism for S100 signalling. Biochem. Cell. Biol. 1998, 76, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Schafer, B.W.; Heizmann, C.W. The S100 family of EF-hand calcium-binding proteins: Functions and pathology. Trends Biochem. Sci. 1996, 21, 134–140. [Google Scholar] [CrossRef]
- Zimmer, D.B.; Cornwall, E.H.; Landar, A.; Song, W. The S100 protein family: History, function, and expression. Brain Res. Bull. 1995, 37, 417–429. [Google Scholar] [CrossRef]
- Kligman, D.; Hilt, D.C. The S100 protein family. Trends Biochem. Sci. 1988, 13, 437–443. [Google Scholar] [CrossRef]
- Heizmann, C.W. S100 proteins: Diagnostic and prognostic biomarkers in laboratory medicine. Biochim. Biophys. Acta Mol. Cell. Res. 2019, 1866, 1197–1206. [Google Scholar] [CrossRef]
- Griffin, W.S.; Stanley, L.C.; Ling, C.; White, L.; MacLeod, V.; Perrot, L.J.; White, C.L., 3rd; Araoz, C. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc. Natl. Acad. Sci. USA 1989, 86, 7611–7615. [Google Scholar] [CrossRef]
- Cross, S.S.; Hamdy, F.C.; Deloulme, J.C.; Rehman, I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology 2005, 46, 256–269. [Google Scholar] [CrossRef]
- Stanley, L.C.; Mrak, R.E.; Woody, R.C.; Perrot, L.J.; Zhang, S.; Marshak, D.R.; Nelson, S.J.; Griffin, W.S. Glial cytokines as neuropathogenic factors in HIV infection: Pathogenic similarities to Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 1994, 53, 231–238. [Google Scholar] [CrossRef]
- Austermann, J.; Spiekermann, C.; Roth, J. S100 proteins in rheumatic diseases. Nat. Rev. Rheumatol. 2018, 14, 528–541. [Google Scholar] [CrossRef]
- Bresnick, A.R. S100 proteins as therapeutic targets. Biophys. Rev. 2018, 10, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Heizmann, C.W. Ca(2+)-binding proteins of the EF-hand superfamily: Diagnostic and prognostic biomarkers and novel therapeutic targets. Methods Mol. Biol. 2019, 1929, 157–186. [Google Scholar] [CrossRef] [PubMed]
- Austermann, J.; Zenker, S.; Roth, J. S100-alarmins: Potential therapeutic targets for arthritis. Expert Opin. Ther. Targets 2017, 21, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Weng, X.; Luecke, H.; Song, I.S.; Kang, D.S.; Kim, S.H.; Huber, R. Crystal structure of human annexin I at 2.5 A resolution. Protein Sci. 1993, 2, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.D.; Moss, S.E. Structural evolution of the annexin supergene family. Trends Genet. 1994, 10, 241–246. [Google Scholar] [CrossRef]
- Freye-Minks, C.; Kretsinger, R.H.; Creutz, C.E. Structural and dynamic changes in human annexin VI induced by a phosphorylation-mimicking mutation, T356D. Biochemistry 2003, 42, 620–630. [Google Scholar] [CrossRef]
- Avila-Sakar, A.J.; Kretsinger, R.H.; Creutz, C.E. Membrane-bound 3D structures reveal the intrinsic flexibility of annexin VI. J. Struct. Biol. 2000, 130, 54–62. [Google Scholar] [CrossRef]
- Shatsky, M.; Nussinov, R.; Wolfson, H.J. A method for simultaneous alignment of multiple protein structures. Proteins 2004, 56, 143–156. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Hofmann, A.; Raguenes-Nicol, C.; Favier-Perron, B.; Mesonero, J.; Huber, R.; Russo-Marie, F.; Lewit-Bentley, A. The annexin A3-membrane interaction is modulated by an N-terminal tryptophan. Biochemistry 2000, 39, 7712–7721. [Google Scholar] [CrossRef]
- Mailliard, W.S.; Haigler, H.T.; Schlaepfer, D.D. Calcium-dependent binding of S100C to the N-terminal domain of annexin I. J. Biol. Chem. 1996, 271, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Seemann, J.; Weber, K.; Gerke, V. Structural requirements for annexin I-S100C complex-formation. Biochem. J. 1996, 319 Pt 1, 123–129. [Google Scholar] [CrossRef]
- Becker, T.; Weber, K.; Johnsson, N. Protein-protein recognition via short amphiphilic helices; a mutational analysis of the binding site of annexin II for p11. EMBO J. 1990, 9, 4207–4213. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.B.; Kraus-Friedmann, N.; Dedman, J.R. Unique calcium-dependent hydrophobic binding proteins: Possible independent mediators of intracellular calcium distinct from calmodulin. J. Cell. Sci. 1984, 72, 121–133. [Google Scholar] [PubMed]
- Glenney, J.R., Jr.; Tack, B.; Powell, M.A. Calpactins: Two distinct Ca++-regulated phospholipid- and actin-binding proteins isolated from lung and placenta. J. Cell. Biol. 1987, 104, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Creutz, C.E.; Zaks, W.J.; Hamman, H.C.; Crane, S.; Martin, W.H.; Gould, K.L.; Oddie, K.M.; Parsons, S.J. Identification of chromaffin granule-binding proteins. Relationship of the chromobindins to calelectrin, synhibin, and the tyrosine kinase substrates p35 and p36. J. Biol. Chem. 1987, 262, 1860–1868. [Google Scholar]
- Flower, R.J. Background and discovery of lipocortins. Agents Actions 1986, 17, 255–262. [Google Scholar] [CrossRef]
- Creutz, C.E.; Pazoles, C.J.; Pollard, H.B. Identification and purification of an adrenal medullary protein (synexin) that causes calcium-dependent aggregation of isolated chromaffin granules. J. Biol. Chem. 1978, 253, 2858–2866. [Google Scholar]
- Crumpton, M.J.; Dedman, J.R. Protein terminology tangle. Nature 1990, 345, 212. [Google Scholar] [CrossRef]
- Monastyrskaya, K.; Babiychuk, E.B.; Hostettler, A.; Rescher, U.; Draeger, A. Annexins as intracellular calcium sensors. Cell Calcium 2007, 41, 207–219. [Google Scholar] [CrossRef]
- Eberhard, D.A.; Karns, L.R.; VandenBerg, S.R.; Creutz, C.E. Control of the nuclear-cytoplasmic partitioning of annexin II by a nuclear export signal and by p11 binding. J. Cell Sci. 2001, 114, 3155–3166. [Google Scholar] [PubMed]
- Merrifield, C.J.; Rescher, U.; Almers, W.; Proust, J.; Gerke, V.; Sechi, A.S.; Moss, S.E. Annexin 2 has an essential role in actin-based macropinocytic rocketing. Curr. Biol. 2001, 11, 1136–1141. [Google Scholar] [CrossRef]
- Rescher, U.; Zobiack, N.; Gerke, V. Intact Ca(2+)-binding sites are required for targeting of annexin 1 to endosomal membranes in living HeLa cells. J. Cell Sci. 2000, 113 Pt 22, 3931–3938. [Google Scholar]
- Langen, R.; Isas, J.M.; Hubbell, W.L.; Haigler, H.T. A transmembrane form of annexin XII detected by site-directed spin labeling. Proc. Natl. Acad. Sci. USA 1998, 95, 14060–14065. [Google Scholar] [CrossRef] [PubMed]
- Langen, R.; Isas, J.M.; Luecke, H.; Haigler, H.T.; Hubbell, W.L. Membrane-mediated assembly of annexins studied by site-directed spin labeling. J. Biol. Chem. 1998, 273, 22453–22457. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, Y.; Mailliard, W.S.; Tranngo, N.; Isas, M.; Luecke, H.; Haigler, H.T.; Hall, J.E. Annexins V and XII alter the properties of planar lipid bilayers seen by conductance probes. J. Gen. Physiol. 2000, 115, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Creutz, C.E.; Moss, S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005, 6, 449–461. [Google Scholar] [CrossRef] [PubMed]
- van Genderen, H.O.; Kenis, H.; Hofstra, L.; Narula, J.; Reutelingsperger, C.P. Extracellular annexin A5: Functions of phosphatidylserine-binding and two-dimensional crystallization. Biochim. Biophys. Acta 2008, 1783, 953–963. [Google Scholar] [CrossRef]
- Hayes, M.J.; Longbottom, R.E.; Evans, M.A.; Moss, S.E. Annexinopathies. Subcell. Biochem. 2007, 45, 1–28. [Google Scholar]
- Bruschi, M.; Petretto, A.; Vaglio, A.; Santucci, L.; Candiano, G.; Ghiggeri, G.M. Annexin A1 and autoimmunity: From basic science to clinical applications. Int. J. Mol. Sci. 2018, 19, 1348. [Google Scholar] [CrossRef]
- Perucci, L.O.; Carneiro, F.S.; Ferreira, C.N.; Sugimoto, M.A.; Soriani, F.M.; Martins, G.G.; Lima, K.M.; Guimaraes, F.L.; Teixeira, A.L.; Dusse, L.M.; et al. Annexin A1 is increased in the plasma of preeclamptic women. PLoS ONE 2015, 10, e0138475. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Zhou, H.; Zhang, Q.; Jin, Y.; Fu, C. Advancements of annexin A1 in inflammation and tumorigenesis. Onco Targets Ther. 2019, 12, 3245–3254. [Google Scholar] [CrossRef] [PubMed]
- Cristante, E.; McArthur, S.; Mauro, C.; Maggioli, E.; Romero, I.A.; Wylezinska-Arridge, M.; Couraud, P.O.; Lopez-Tremoleda, J.; Christian, H.C.; Weksler, B.B.; et al. Identification of an essential endogenous regulator of blood-brain barrier integrity, and its pathological and therapeutic implications. Proc. Natl. Acad. Sci. USA 2013, 110, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.H.; Solito, E. Annexin A1: Uncovering the many talents of an old protein. Int. J. Mol. Sci. 2018, 19, 1045. [Google Scholar] [CrossRef]
- Hitchcock, J.K.; Katz, A.A.; Schafer, G. Dynamic reciprocity: The role of annexin A2 in tissue integrity. J. Cell Commun. Signal. 2014, 8, 125–133. [Google Scholar] [CrossRef]
- Canas, F.; Simonin, L.; Couturaud, F.; Renaudineau, Y. Annexin A2 autoantibodies in thrombosis and autoimmune diseases. Thromb. Res. 2015, 135, 226–230. [Google Scholar] [CrossRef]
- Sharma, M.C. Annexin A2 (ANX A2): An emerging biomarker and potential therapeutic target for aggressive cancers. Int. J. Cancer 2019, 144, 2074–2081. [Google Scholar] [CrossRef]
- Xu, X.H.; Pan, W.; Kang, L.H.; Feng, H.; Song, Y.Q. Association of annexin A2 with cancer development (Review). Oncol. Rep. 2015, 33, 2121–2128. [Google Scholar] [CrossRef]
- Li, J.; Zhou, T.; Liu, L.; Ju, Y.C.; Chen, Y.T.; Tan, Z.R.; Wang, J. The regulatory role of Annexin 3 in a nude mouse bearing a subcutaneous xenograft of MDA-MB-231 human breast carcinoma. Pathol. Res. Pract. 2018, 214, 1719–1725. [Google Scholar] [CrossRef]
- Tong, M.; Fung, T.M.; Luk, S.T.; Ng, K.Y.; Lee, T.K.; Lin, C.H.; Yam, J.W.; Chan, K.W.; Ng, F.; Zheng, B.J.; et al. ANXA3/JNK signaling promotes self-renewal and tumor growth, and its blockade provides a therapeutic target for hepatocellular carcinoma. Stem. Cell Rep. 2015, 5, 45–59. [Google Scholar] [CrossRef]
- Pan, Q.Z.; Pan, K.; Wang, Q.J.; Weng, D.S.; Zhao, J.J.; Zheng, H.X.; Zhang, X.F.; Jiang, S.S.; Lv, L.; Tang, Y.; et al. Annexin A3 as a potential target for immunotherapy of liver cancer stem-like cells. Stem Cells 2015, 33, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.Z.; Pan, K.; Weng, D.S.; Zhao, J.J.; Zhang, X.F.; Wang, D.D.; Lv, L.; Jiang, S.S.; Zheng, H.X.; Xia, J.C. Annexin A3 promotes tumorigenesis and resistance to chemotherapy in hepatocellular carcinoma. Mol. Carcinog. 2015, 54, 598–607. [Google Scholar] [CrossRef] [PubMed]
- de la Cuesta, F.; Alvarez-Llamas, G.; Maroto, A.S.; Donado, A.; Zubiri, I.; Posada, M.; Padial, L.R.; Pinto, A.G.; Barderas, M.G.; Vivanco, F. A proteomic focus on the alterations occurring at the human atherosclerotic coronary intima. Mol. Cell. Proteom. 2011, 10, M110-003517. [Google Scholar] [CrossRef] [PubMed]
- Pogosian, G.G.; Mikaelian, M.V.; Avagian, A.; Gasparian, V.K. The annexin 5 in serums of pregnant women and patients with particular types of cancer. Klin. Lab. Diagn. 2014, 4, 14–17. [Google Scholar]
- Emanuel, V.L.; Mnuskina, M.M.; Smirnov, A.V.; Panina, I.; Rumiantsev, A.; Vasina, E.; Vasina, L.B. Annexin-5 as a biochemical marker of early vascular disorders under chronic disease of kidneys. Klin. Lab. Diagn. 2013, 4, 9–10. [Google Scholar]
- Burkhard, F.C.; Monastyrskaya, K.; Studer, U.E.; Draeger, A. Smooth muscle membrane organization in the normal and dysfunctional human urinary bladder: A structural analysis. Neurourol. Urodyn. 2005, 24, 128–135. [Google Scholar] [CrossRef]
- Srivastava, M.; Montagna, C.; Leighton, X.; Glasman, M.; Naga, S.; Eidelman, O.; Ried, T.; Pollard, H.B. Haploinsufficiency of Anx7 tumor suppressor gene and consequent genomic instability promotes tumorigenesis in the Anx7(+/-) mouse. Proc. Natl. Acad. Sci. USA 2003, 100, 14287–14292. [Google Scholar] [CrossRef]
- Lang, F.; Qadri, S.M. Mechanisms and significance of eryptosis, the suicidal death of erythrocytes. Blood Purif. 2012, 33, 125–130. [Google Scholar] [CrossRef]
- Rossetti, S.; Bshara, W.; Reiners, J.A.; Corlazzoli, F.; Miller, A.; Sacchi, N. Harnessing 3D models of mammary epithelial morphogenesis: An off the beaten path approach to identify candidate biomarkers of early stage breast cancer. Cancer Lett. 2016, 380, 375–383. [Google Scholar] [CrossRef]
- Calderon-Gonzalez, K.G.; Valero Rustarazo, M.L.; Labra-Barrios, M.L.; Bazan-Mendez, C.I.; Tavera-Tapia, A.; Herrera-Aguirre, M.E.; Sanchez del Pino, M.M.; Gallegos-Perez, J.L.; Gonzalez-Marquez, H.; Hernandez-Hernandez, J.M.; et al. Determination of the protein expression profiles of breast cancer cell lines by quantitative proteomics using iTRAQ labelling and tandem mass spectrometry. J. Proteom. 2015, 124, 50–78. [Google Scholar] [CrossRef]
- Salom, C.; Alvarez-Teijeiro, S.; Fernandez, M.P.; Morgan, R.O.; Allonca, E.; Vallina, A.; Lorz, C.; de Villalain, L.; Fernandez-Garcia, M.S.; Rodrigo, J.P.; et al. Frequent alteration of annexin A9 and A10 in HPV-negative head and neck squamous cell carcinomas: Correlation with the histopathological differentiation grade. J. Clin. Med. 2019, 8, 229. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guo, C.; Liu, S.; Qi, H.; Yin, Y.; Liang, R.; Sun, M.Z.; Greenaway, F.T. Annexin A11 in disease. Clin. Chim. Acta 2014, 431, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Benjachat, T.; Tongyoo, P.; Tantivitayakul, P.; Somparn, P.; Hirankarn, N.; Prom-On, S.; Pisitkun, P.; Leelahavanichkul, A.; Avihingsanon, Y.; Townamchai, N. Biomarkers for refractory lupus nephritis: A microarray study of kidney tissue. Int. J. Mol. Sci. 2015, 16, 14276–14290. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The string database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Rintala-Dempsey, A.C.; Rezvanpour, A.; Shaw, G.S. S100-annexin complexes--structural insights. FEBS J. 2008, 275, 4956–4966. [Google Scholar] [CrossRef] [PubMed]
- Lewit-Bentley, A.; Rety, S.; Sopkova-de Oliveira Santos, J.; Gerke, V. S100-annexin complexes: Some insights from structural studies. Cell Biol. Int. 2000, 24, 799–802. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002, 12, 54–60. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002, 323, 573–584. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef]
- Fink, A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 2005, 15, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your ID: Intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005, 18, 343–384. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Dunker, A.K. Biochemistry. Controlled chaos. Science 2008, 322, 1340–1341. [Google Scholar] [CrossRef]
- Tompa, P.; Fuxreiter, M.; Oldfield, C.J.; Simon, I.; Dunker, A.K.; Uversky, V.N. Close encounters of the third kind: Disordered domains and the interactions of proteins. Bioessays 2009, 31, 328–335. [Google Scholar] [CrossRef]
- Dunker, A.K.; Uversky, V.N. Signal transduction via unstructured protein conduits. Nat. Chem. Biol. 2008, 4, 229–230. [Google Scholar] [CrossRef]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Meng, J.; Yang, J.Y.; Yang, M.Q.; Uversky, V.N.; Dunker, A.K. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genom. 2008, 9 (Suppl. 1), S1. [Google Scholar] [CrossRef]
- Bertini, I.; Das Gupta, S.; Hu, X.; Karavelas, T.; Luchinat, C.; Parigi, G.; Yuan, J. Solution structure and dynamics of S100A5 in the apo and Ca2+-bound states. J. Biol. Inorg. Chem. 2009, 14, 1097–1107. [Google Scholar] [CrossRef]
- Malik, S.; Revington, M.; Smith, S.P.; Shaw, G.S. Analysis of the structure of human apo-S100B at low temperature indicates a unimodal conformational distribution is adopted by calcium-free S100 proteins. Proteins 2008, 73, 28–42. [Google Scholar] [CrossRef]
- Skelton, N.J.; Kordel, J.; Chazin, W.J. Determination of the solution structure of Apo calbindin D9k by NMR spectroscopy. J. Mol. Biol. 1995, 249, 441–462. [Google Scholar] [CrossRef] [PubMed]
- Dutta, K.; Cox, C.J.; Basavappa, R.; Pascal, S.M. 15N relaxation studies of Apo-Mts1: A dynamic S100 protein. Biochemistry 2008, 47, 7637–7647. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Bioinform. 2005, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Ismailov, R.G.; Xue, B.; Denesyuk, A.I.; Uversky, V.N.; Permyakov, E.A. Intrinsic disorder in S100 proteins. Mol. Biosyst. 2011, 7, 2164–2180. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins Struct. Funct. Bioinform. 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Hu, N.J.; Bradshaw, J.; Lauter, H.; Buckingham, J.; Solito, E.; Hofmann, A. Membrane-induced folding and structure of membrane-bound annexin A1 N-terminal peptides: Implications for annexin-induced membrane aggregation. Biophys. J. 2008, 94, 1773–1781. [Google Scholar] [CrossRef]
- Yoon, M.K.; Park, S.H.; Won, H.S.; Na, D.S.; Lee, B.J. Solution structure and membrane-binding property of the N-terminal tail domain of human annexin I. FEBS Lett. 2000, 484, 241–245. [Google Scholar] [CrossRef]
- McCulloch, K.M.; Yamakawa, I.; Shifrin, D.A., Jr.; McConnell, R.E.; Foegeding, N.J.; Singh, P.K.; Mao, S.; Tyska, M.J.; Iverson, T.M. An alternative N-terminal fold of the intestine-specific annexin A13a induces dimerization and regulates membrane-binding. J. Biol. Chem. 2019, 294, 3454–3463. [Google Scholar] [CrossRef]
- Oates, M.E.; Romero, P.; Ishida, T.; Ghalwash, M.; Mizianty, M.J.; Xue, B.; Dosztanyi, Z.; Uversky, V.N.; Obradovic, Z.; Kurgan, L.; et al. D(2)P(2): Database of disordered protein predictions. Nucleic Acids Res. 2013, 41, D508–D516. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [PubMed]
- Walsh, I.; Martin, A.J.; Di Domenico, T.; Tosatto, S.C. ESpritz: Accurate and fast prediction of protein disorder. Bioinformatics 2012, 28, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Oldfield, C.J.; Meng, J.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 2007, 46, 13468–13477. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454–12470. [Google Scholar] [CrossRef]
- Meszaros, B.; Simon, I.; Dosztanyi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Meszaros, B.; Simon, I. ANCHOR: Web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009, 25, 2745–2746. [Google Scholar] [CrossRef]
- Arcuri, C.; Giambanco, I.; Bianchi, R.; Donato, R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience 2002, 109, 371–388. [Google Scholar] [CrossRef]
- Garbuglia, M.; Verzini, M.; Donato, R. Annexin VI binds S100A1 and S100B and blocks the ability of S100A1 and S100B to inhibit desmin and GFAP assemblies into intermediate filaments. Cell Calcium 1998, 24, 177–191. [Google Scholar] [CrossRef]
- Garbuglia, M.; Verzini, M.; Hofmann, A.; Huber, R.; Donato, R. S100A1 and S100B interactions with annexins. Biochim. Biophys. Acta 2000, 1498, 192–206. [Google Scholar] [CrossRef]
- Semov, A.; Moreno, M.J.; Onichtchenko, A.; Abulrob, A.; Ball, M.; Ekiel, I.; Pietrzynski, G.; Stanimirovic, D.; Alakhov, V. Metastasis-associated protein S100A4 induces angiogenesis through interaction with annexin II and accelerated plasmin formation. J. Biol. Chem. 2005, 280, 20833–20841. [Google Scholar] [CrossRef] [PubMed]
- Ecsedi, P.; Kiss, B.; Gogl, G.; Radnai, L.; Buday, L.; Koprivanacz, K.; Liliom, K.; Leveles, I.; Vertessy, B.; Jeszenoi, N.; et al. Regulation of the equilibrium between closed and open conformations of annexin A2 by N-terminal phosphorylation and S100A4-binding. Structure 2017, 25, 1195–1207.e5. [Google Scholar] [CrossRef] [PubMed]
- Sopkova-de Oliveira Santos, J.; Oling, F.K.; Rety, S.; Brisson, A.; Smith, J.C.; Lewit-Bentley, A. S100 protein-annexin interactions: A model of the (Anx2-p11)(2) heterotetramer complex. Biochim. Biophys. Acta 2000, 1498, 181–191. [Google Scholar] [CrossRef]
- Glenney, J.R., Jr.; Boudreau, M.; Galyean, R.; Hunter, T.; Tack, B. Association of the S-100-related calpactin I light chain with the NH2-terminal tail of the 36-kDa heavy chain. J. Biol. Chem. 1986, 261, 10485–10488. [Google Scholar] [PubMed]
- Kube, E.; Becker, T.; Weber, K.; Gerke, V. Protein-protein interaction studied by site-directed mutagenesis. Characterization of the annexin II-binding site on p11, a member of the S100 protein family. J. Biol. Chem. 1992, 267, 14175–14182. [Google Scholar]
- Jost, M.; Gerke, V. Mapping of a regulatory important site for protein kinase C phosphorylation in the N-terminal domain of annexin II. Biochim. Biophys. Acta 1996, 1313, 283–289. [Google Scholar] [CrossRef]
- Johnsson, N.; Marriott, G.; Weber, K. P36, the major cytoplasmic substrate of src tyrosine protein kinase, binds to its p11 regulatory subunit via a short amino-terminal amphiphatic helix. EMBO J. 1988, 7, 2435–2442. [Google Scholar] [CrossRef]
- Lambert, O.; Gerke, V.; Bader, M.F.; Porte, F.; Brisson, A. Structural analysis of junctions formed between lipid membranes and several annexins by cryo-electron microscopy. J. Mol. Biol. 1997, 272, 42–55. [Google Scholar] [CrossRef]
- Menke, M.; Ross, M.; Gerke, V.; Steinem, C. The molecular arrangement of membrane-bound annexin A2-S100A10 tetramer as revealed by scanning force microscopy. Chembiochem 2004, 5, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Cesarman-Maus, G.; Hajjar, K.A. Molecular mechanisms of fibrinolysis. Br. J. Haematol. 2005, 129, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Tinel, N.; Terrenoire, C.; Romey, G.; Lazdunski, M.; Borsotto, M. P11, an annexin II subunit, an auxiliary protein associated with the background K+ channel, TASK-1. EMBO J. 2002, 21, 4439–4448. [Google Scholar] [CrossRef] [PubMed]
- Renigunta, V.; Yuan, H.; Zuzarte, M.; Rinne, S.; Koch, A.; Wischmeyer, E.; Schlichthorl, G.; Gao, Y.; Karschin, A.; Jacob, R.; et al. The retention factor p11 confers an endoplasmic reticulum-localization signal to the potassium channel TASK-1. Traffic 2006, 7, 168–181. [Google Scholar] [CrossRef]
- Svenningsson, P.; Chergui, K.; Rachleff, I.; Flajolet, M.; Zhang, X.; El Yacoubi, M.; Vaugeois, J.M.; Nomikos, G.G.; Greengard, P. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science 2006, 311, 77–80. [Google Scholar] [CrossRef]
- Okuse, K.; Malik-Hall, M.; Baker, M.D.; Poon, W.Y.; Kong, H.; Chao, M.V.; Wood, J.N. Annexin II light chain regulates sensory neuron-specific sodium channel expression. Nature 2002, 417, 653–656. [Google Scholar] [CrossRef]
- van de Graaf, S.F.; Hoenderop, J.G.; Gkika, D.; Lamers, D.; Prenen, J.; Rescher, U.; Gerke, V.; Staub, O.; Nilius, B.; Bindels, R.J. Functional expression of the epithelial Ca(2+) channels (TRPV5 and TRPV6) requires association of the S100A10-annexin 2 complex. EMBO J. 2003, 22, 1478–1487. [Google Scholar] [CrossRef]
- Hirschhorn, R.R.; Aller, P.; Yuan, Z.A.; Gibson, C.W.; Baserga, R. Cell-cycle-specific cDNAs from mammalian cells temperature sensitive for growth. Proc. Natl. Acad. Sci. USA 1984, 81, 6004–6008. [Google Scholar] [CrossRef]
- Tomas, A.; Moss, S.E. Calcium- and cell cycle-dependent association of annexin 11 with the nuclear envelope. J. Biol. Chem. 2003, 278, 20210–20216. [Google Scholar] [CrossRef]
- Tokumitsu, H.; Mizutani, A.; Minami, H.; Kobayashi, R.; Hidaka, H. A calcyclin-associated protein is a newly identified member of the Ca2+/phospholipid-binding proteins, annexin family. J. Biol. Chem. 1992, 267, 8919–8924. [Google Scholar] [PubMed]
- Williams, L.H.; McClive, P.J.; Van Den Bergen, J.A.; Sinclair, A.H. Annexin XI co-localises with calcyclin in proliferating cells of the embryonic mouse testis. Dev. Dyn. 2005, 234, 432–437. [Google Scholar] [CrossRef]
- Seemann, J.; Weber, K.; Gerke, V. Annexin I targets S100C to early endosomes. FEBS Lett. 1997, 413, 185–190. [Google Scholar] [CrossRef]
- Robinson, N.A.; Lapic, S.; Welter, J.F.; Eckert, R.L. S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J. Biol. Chem. 1997, 272, 12035–12046. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsic disorder-based protein interactions and their modulators. Curr. Pharm. Des. 2013, 19, 4191–4213. [Google Scholar] [CrossRef] [PubMed]
- Jakob, U.; Kriwacki, R.; Uversky, V.N. Conditionally and transiently disordered proteins: Awakening cryptic disorder to regulate protein function. Chem. Rev. 2014, 114, 6779–6805. [Google Scholar] [CrossRef]
- Uversky, V.N. P53 proteoforms and Intrinsic disorder: An illustration of the protein structure-function continuum concept. Int. J. Mol. Sci. 2016, 17, 1874. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L.; Consortium for Top Down, P. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Schluter, H.; Apweiler, R.; Holzhutter, H.G.; Jungblut, P.R. Finding one’s way in proteomics: A protein species nomenclature. Chem. Cent. J. 2009, 3, 11. [Google Scholar] [CrossRef]
- Consortium, T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Farrah, T.; Deutsch, E.W.; Omenn, G.S.; Sun, Z.; Watts, J.D.; Yamamoto, T.; Shteynberg, D.; Harris, M.M.; Moritz, R.L. State of the human proteome in 2013 as viewed through peptideatlas: Comparing the kidney, urine, and plasma proteomes for the biology- and disease-driven human proteome project. J. Proteome Res. 2014, 13, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Farrah, T.; Deutsch, E.W.; Hoopmann, M.R.; Hallows, J.L.; Sun, Z.; Huang, C.Y.; Moritz, R.L. The state of the human proteome in 2012 as viewed through peptideatlas. J. Proteome Res. 2013, 12, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Reddy, P.J.; Ray, S.; Srivastava, S. The quest of the human proteome and the missing proteins: Digging deeper. OMICS J. Integr. Biol. 2015, 19, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef]
- Uversky, V.N. (Intrinsically disordered) splice variants in the proteome: Implications for novel drug discovery. Genes Genom. 2016, 38, 577–594. [Google Scholar] [CrossRef]
- Darling, A.L.; Uversky, V.N. Intrinsic disorder and posttranslational modifications: The darker side of the biological dark matter. Front. Genet. 2018, 9, 158. [Google Scholar] [CrossRef]
- Niklas, K.J.; Bondos, S.E.; Dunker, A.K.; Newman, S.A. Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front. Cell Dev. Biol. 2015, 3, 8. [Google Scholar] [CrossRef]
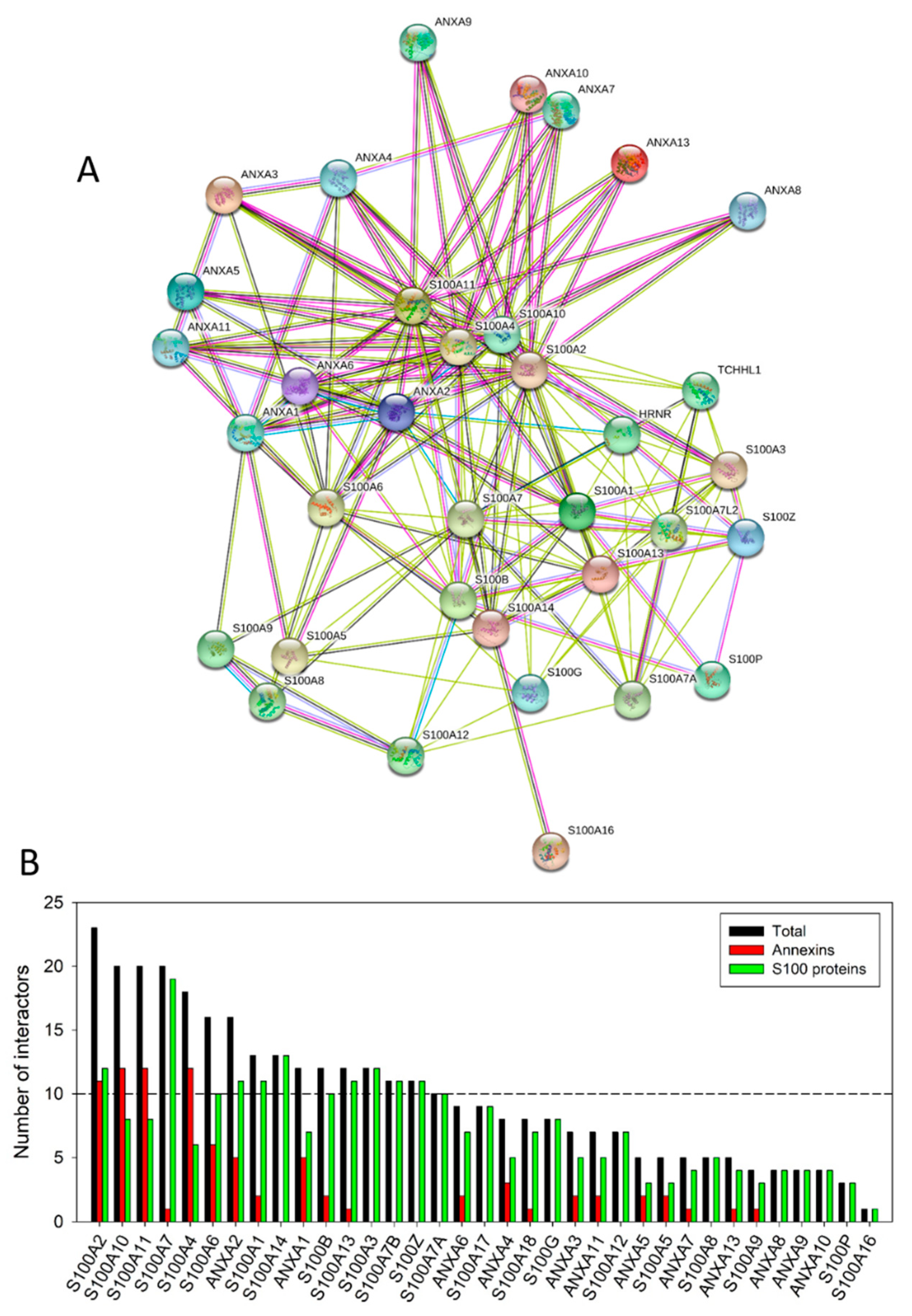
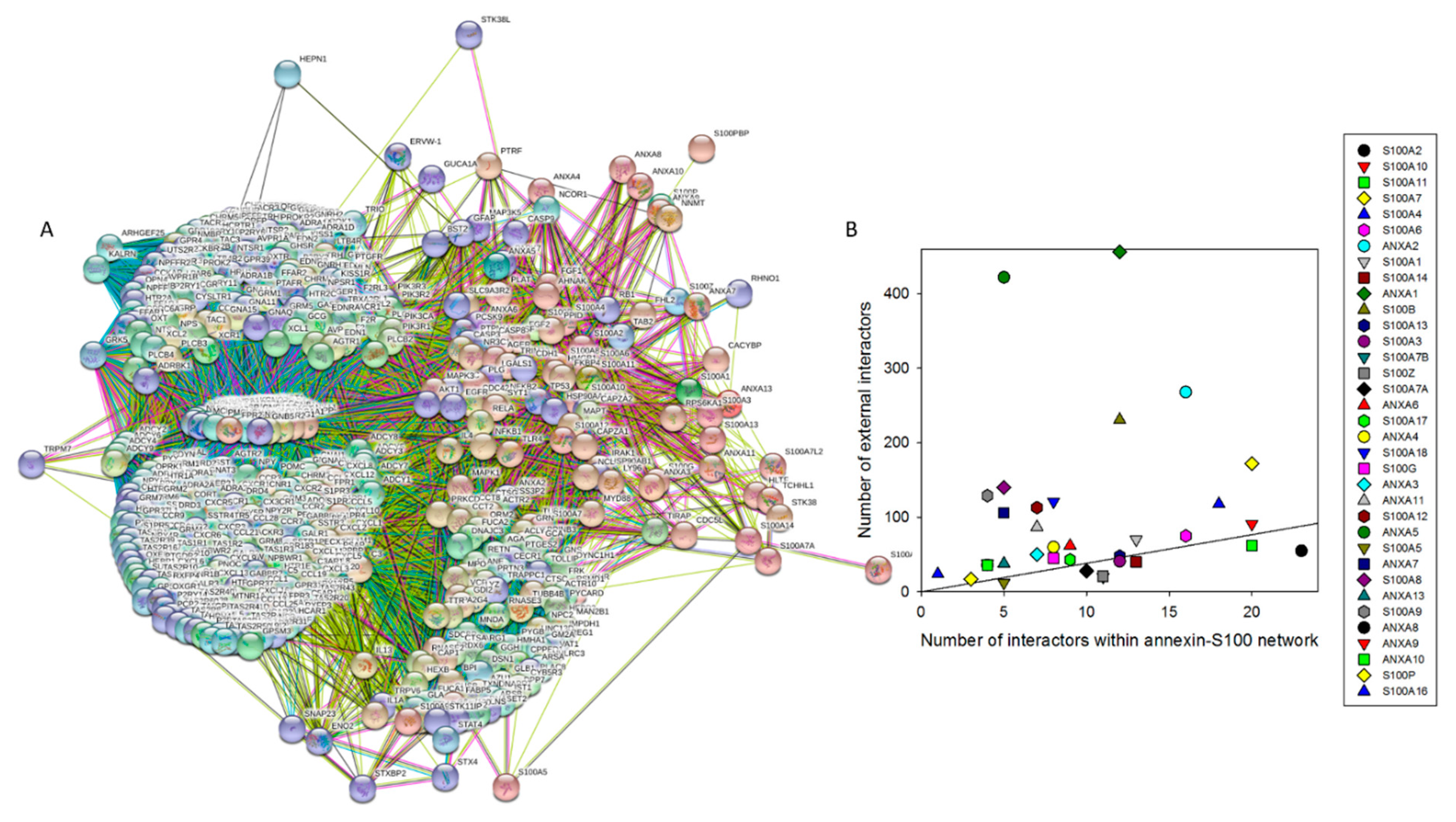
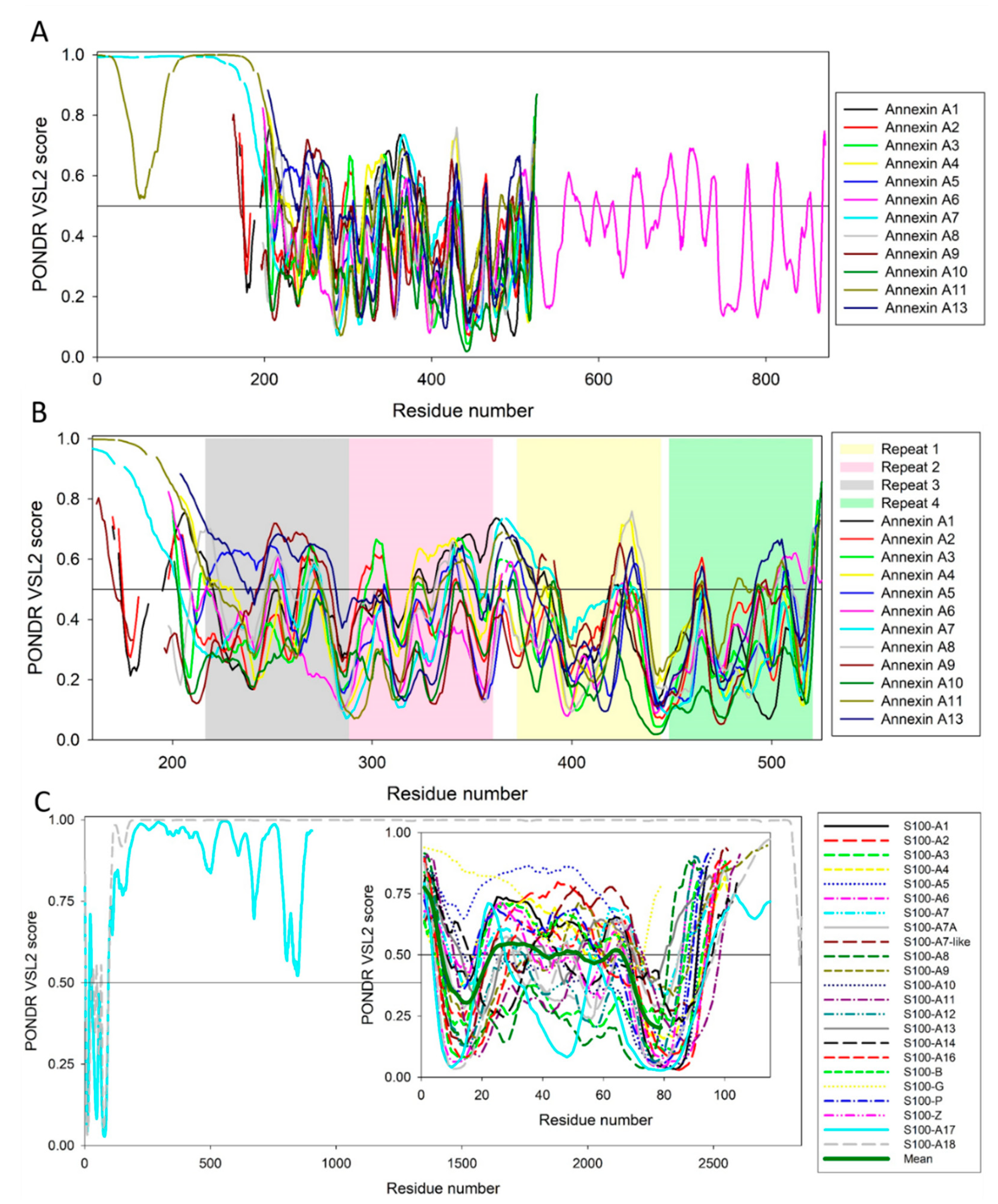
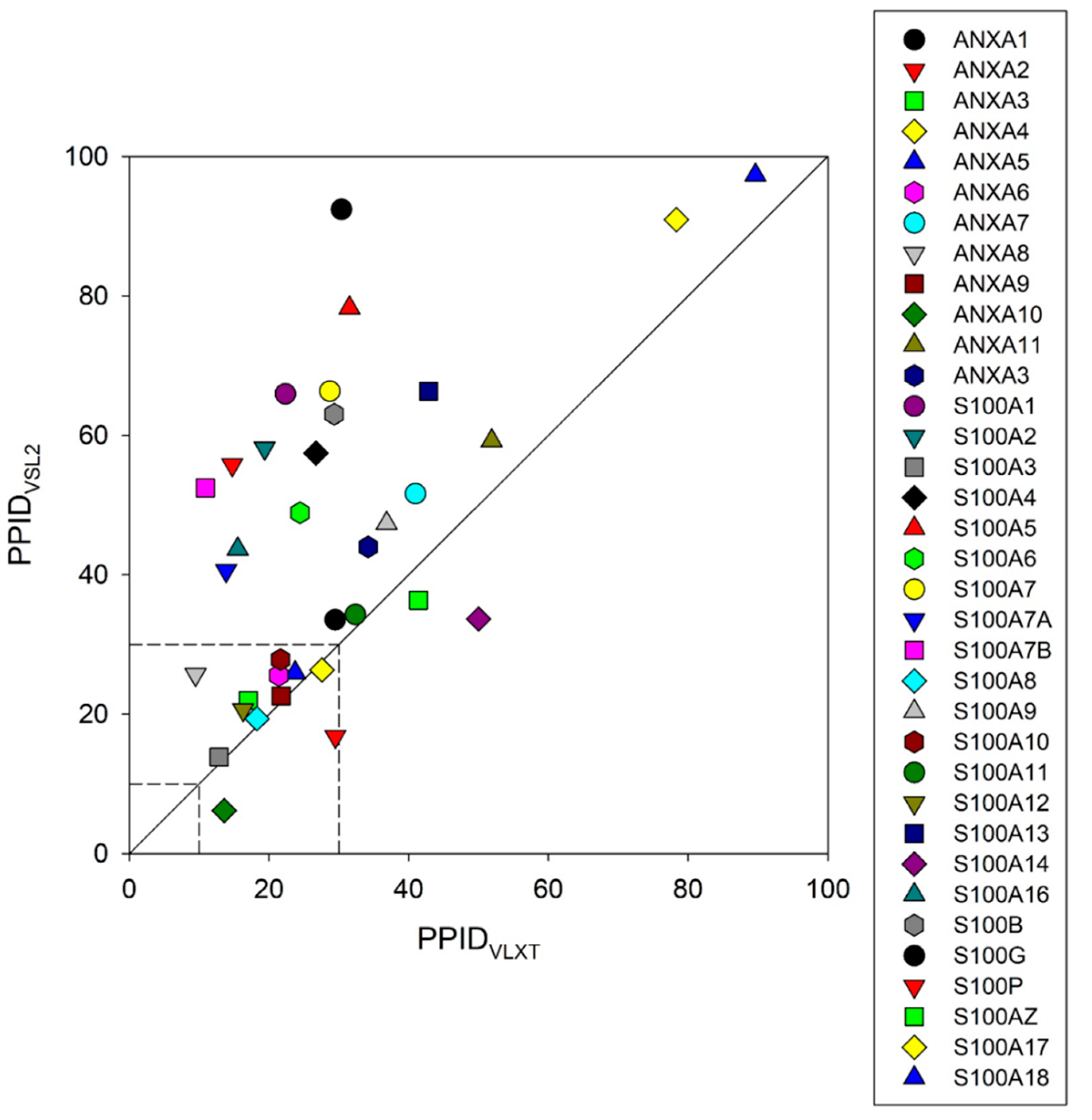

| Protein Name | UniProt ID | NIntAct | NSTRING (0.9) | NSTRING (0.7) | NSTRING (0.4) |
|---|---|---|---|---|---|
| Annexins | |||||
| ANXA1 | P04083 | 89 | 354 | 362 | 456 |
| ANXA2 | P07355 | 102 | 99 | 119 | 268 |
| ANXA3 | P12429 | 7 | 0 | 9 | 50 |
| ANXA4 | P09525 | 11 | 0 | 2 | 60 |
| ANXA5 | P08758 | 52 | 3 | 37 | 422 |
| ANXA6 | P08133 | 24 | 6 | 10 | 62 |
| ANXA7 | P20073 | 122 | 2 | 20 | 106 |
| ANXA8 | P13928 | 13 | 0 | 4 | 36 |
| ANXA9 | O76027 | 5 | 0 | 2 | 38 |
| ANXA10 | Q9UJ72 | 3 | 0 | 3 | 36 |
| ANXA11 | P50995 | 27 | 0 | 12 | 87 |
| ANXA13 | P27216 | 0 | 0 | 5 | 38 |
| S100 proteins | |||||
| S100A1 | P23297 | 16 | 5 | 21 | 70 |
| S100A2 | P29034 | 21 | 0 | 9 | 55 |
| S100A3 | P33764 | 20 | 2 | 8 | 41 |
| S100A4 | P26447 | 44 | 2 | 13 | 118 |
| S100A5 | P33763 | 2 | 0 | 0 | 12 |
| S100A6 | P06703 | 27 | 3 | 13 | 75 |
| S100A7 | P31151 | 41 | 86 | 94 | 172 |
| S100A7B | Q86SG5 | 8 | 0 | 3 | 20 |
| S100A7A | Q5SY68 | 4 | 0 | 0 | 28 |
| S100A8 | P05109 | 59 | 8 | 19 | 140 |
| S100A9 | P06702 | 52 | 8 | 15 | 129 |
| S100A10 | P60903 | 50 | 10 | 20 | 91 |
| S100A11 | P31949 | 15 | 3 | 5 | 62 |
| S100A12 | P80511 | 2 | 15 | 16 | 113 |
| S100A13 | Q99584 | 18 | 4 | 6 | 48 |
| S100A14 | Q9HCY8 | 15 | 0 | 4 | 40 |
| S100A16 | Q96FQ6 | 12 | 0 | 10 | 24 |
| S100B | P04271 | 30 | 19 | 51 | 231 |
| S100Z | P29377 | 0 | 0 | 6 | 21 |
| S100G | P25815 | 20 | 0 | 5 | 45 |
| S100P | Q8WXG8 | 8 | 3 | 7 | 17 |
| S100A17 | Q5QJ38 | 0 | 0 | 6 | 43 |
| S100A18 | Q86YZ3 | 32 | 86 | 87 | 121 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weisz, J.; Uversky, V.N. Zooming into the Dark Side of Human Annexin-S100 Complexes: Dynamic Alliance of Flexible Partners. Int. J. Mol. Sci. 2020, 21, 5879. https://doi.org/10.3390/ijms21165879
Weisz J, Uversky VN. Zooming into the Dark Side of Human Annexin-S100 Complexes: Dynamic Alliance of Flexible Partners. International Journal of Molecular Sciences. 2020; 21(16):5879. https://doi.org/10.3390/ijms21165879
Chicago/Turabian StyleWeisz, Judith, and Vladimir N. Uversky. 2020. "Zooming into the Dark Side of Human Annexin-S100 Complexes: Dynamic Alliance of Flexible Partners" International Journal of Molecular Sciences 21, no. 16: 5879. https://doi.org/10.3390/ijms21165879
APA StyleWeisz, J., & Uversky, V. N. (2020). Zooming into the Dark Side of Human Annexin-S100 Complexes: Dynamic Alliance of Flexible Partners. International Journal of Molecular Sciences, 21(16), 5879. https://doi.org/10.3390/ijms21165879






