The Study of Protein–Cyclitol Interactions
Abstract
:1. Introduction
2. Results and Discussion
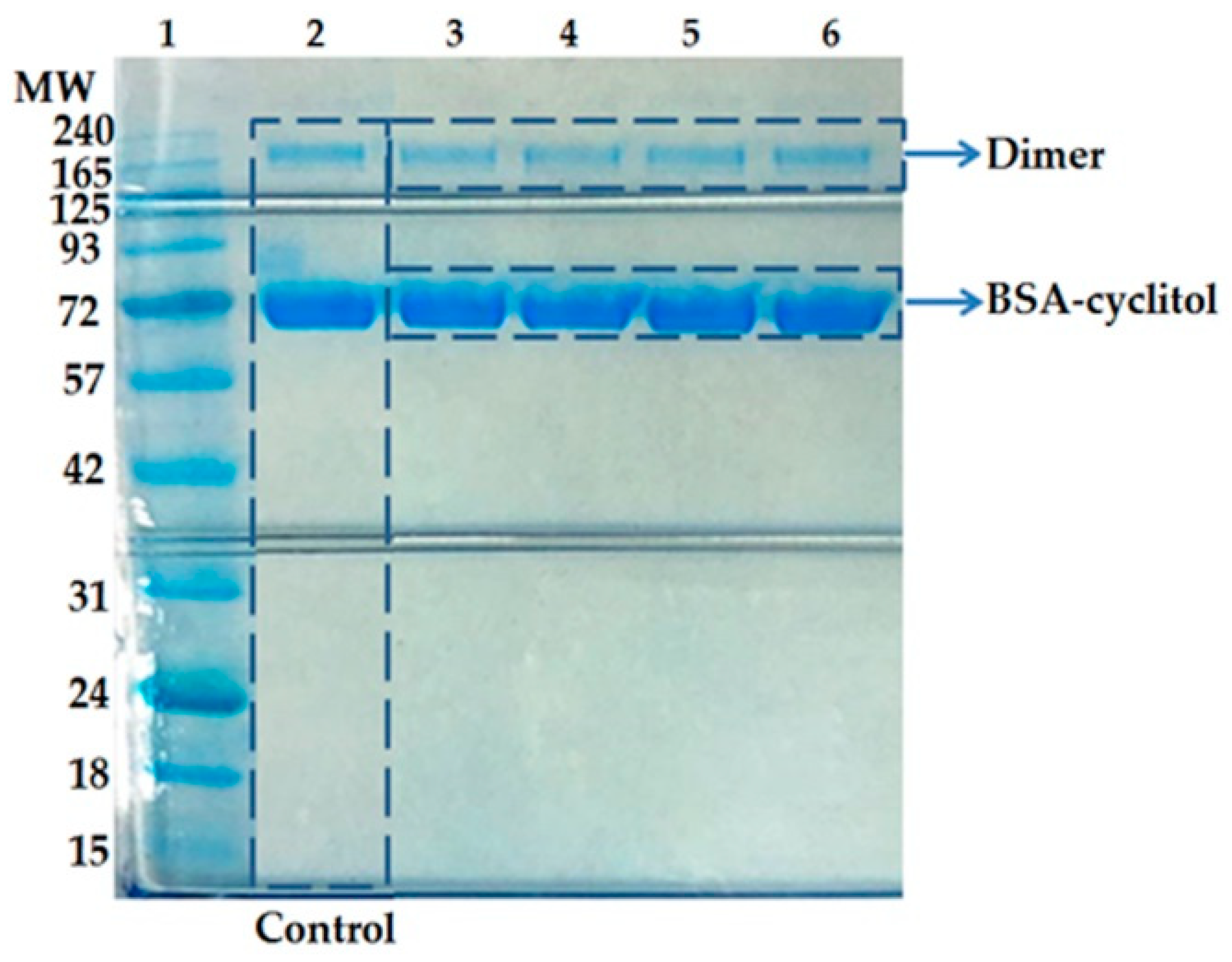
2.1. SDS-PAGE
2.2. Kinetic Modeling of BSA–Cyclitol Interactions
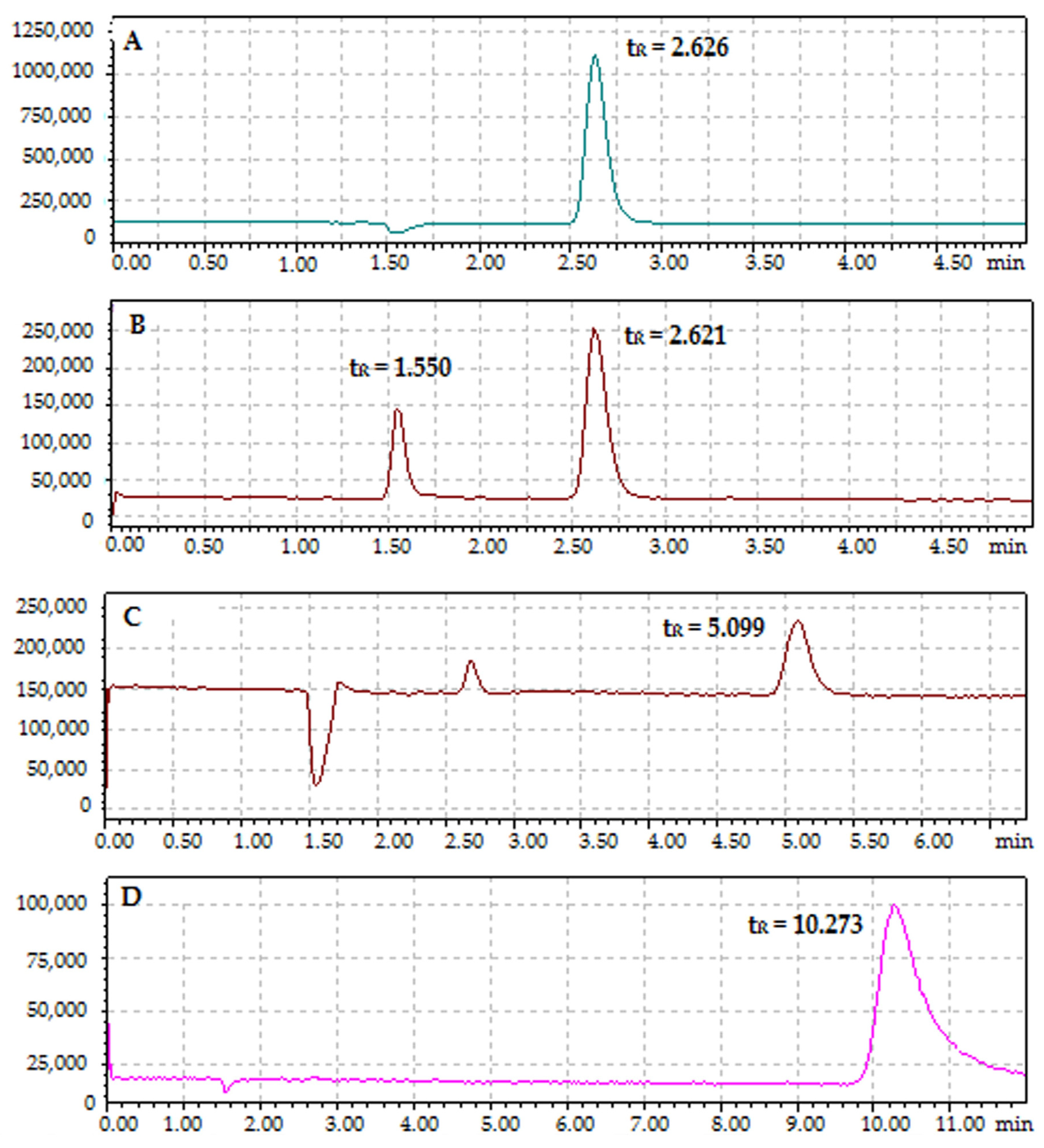
2.3. HPLC-MS/MS Analysis
2.3.1. Analysis of Protein–Cyclitol Interactions
2.3.2. Indication of Unbounded Cyclitols
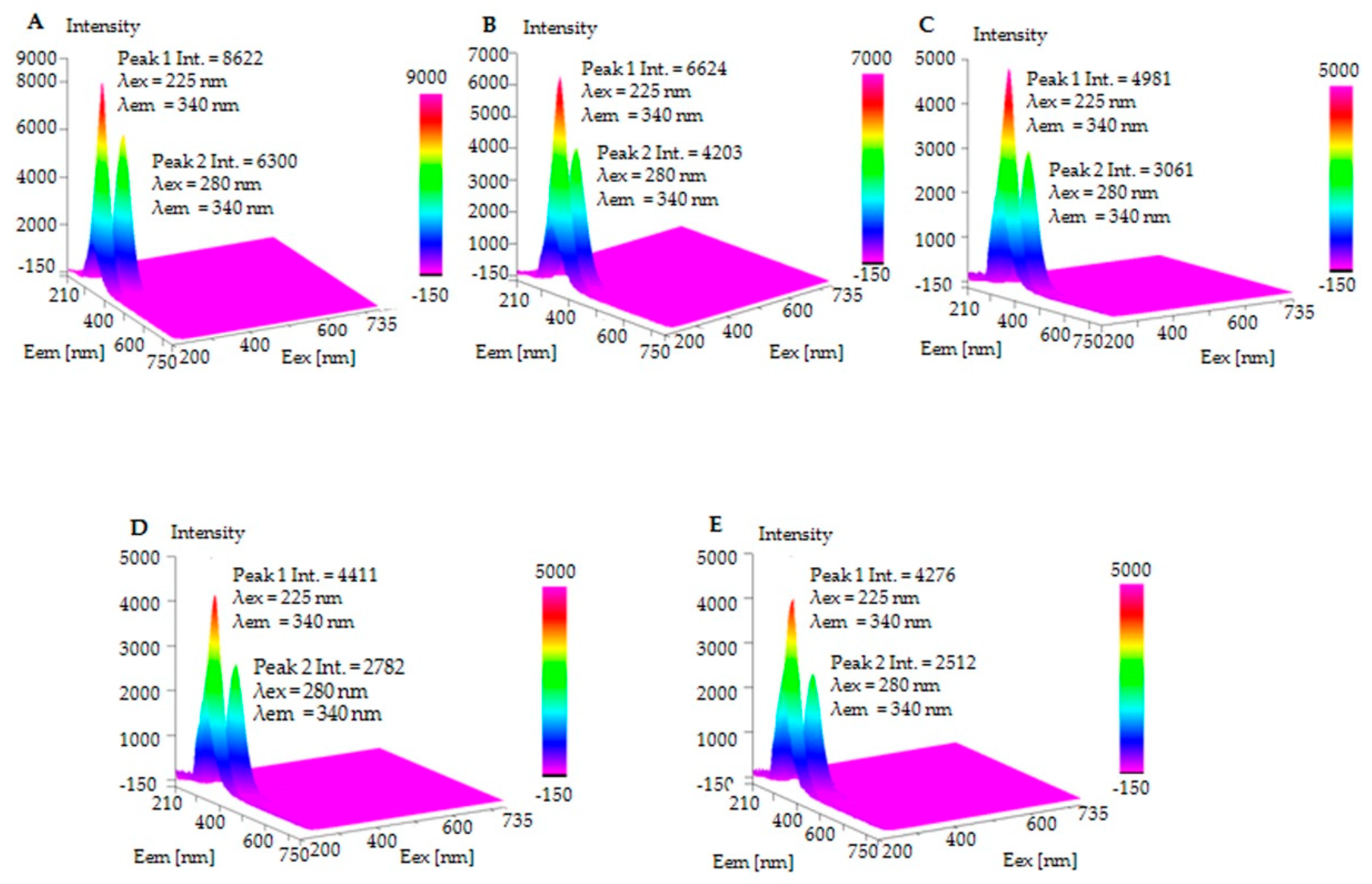
2.4. Fluorescence Spectroscopy
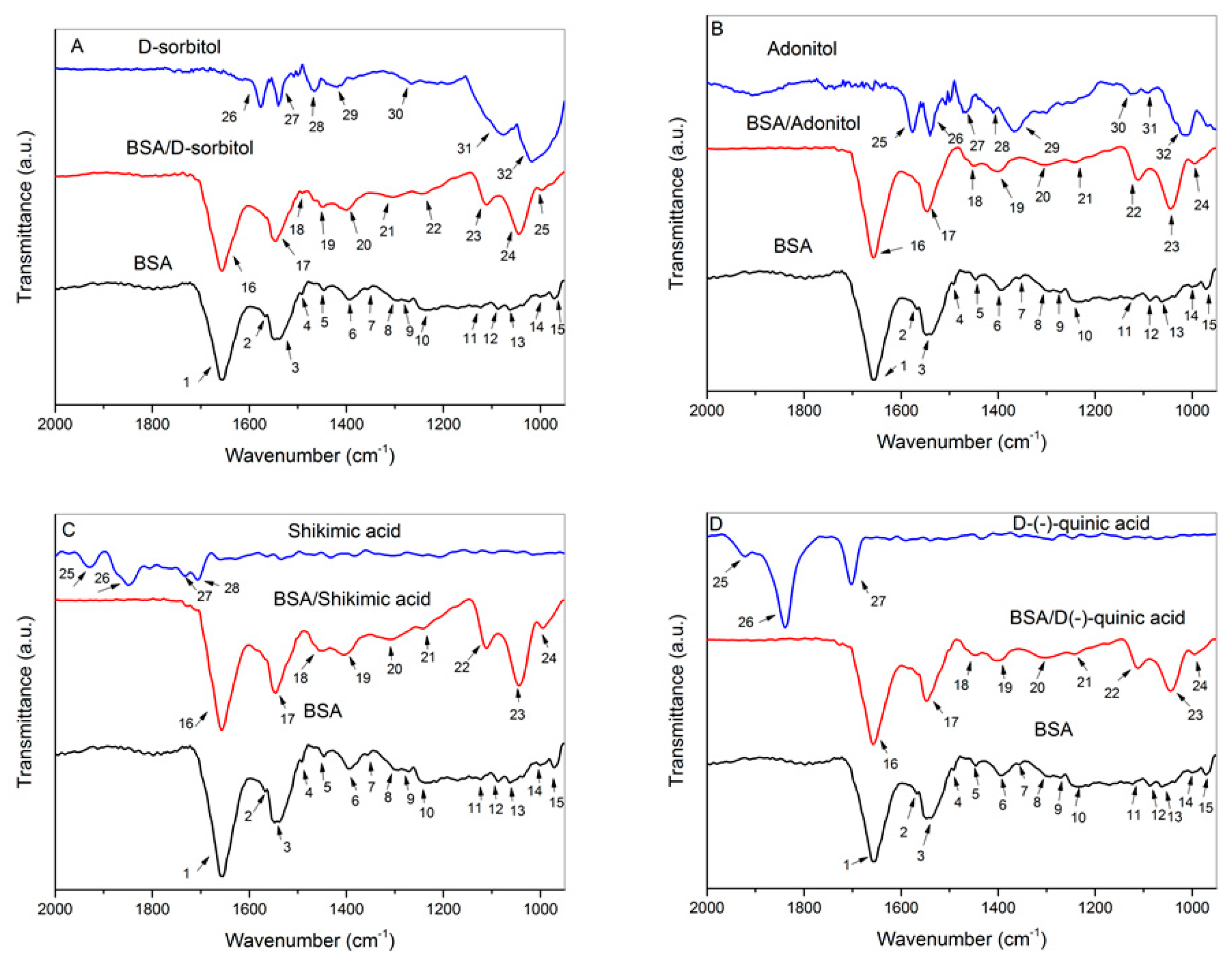
2.5. FTIR Analysis
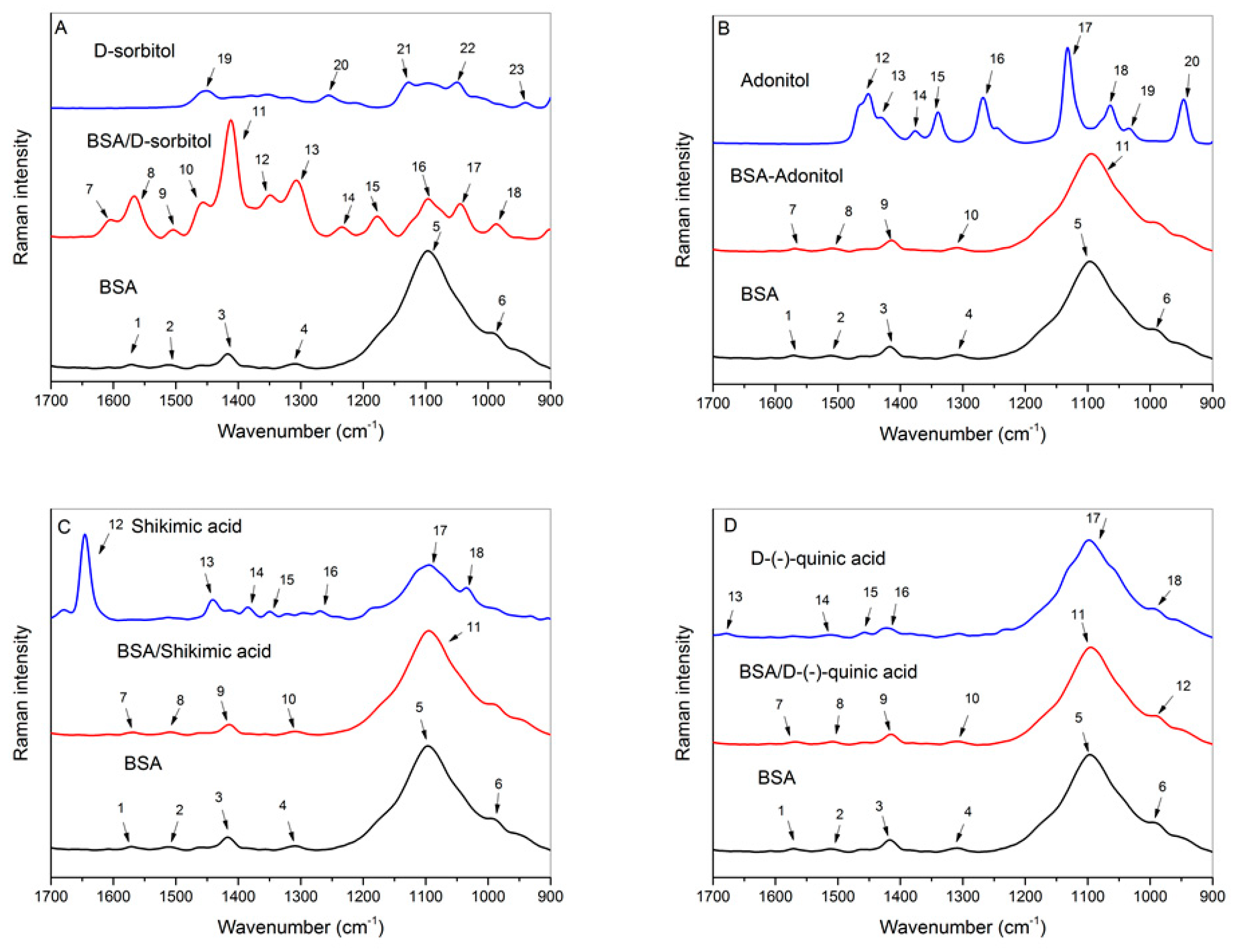
2.6. Raman Spectroscopy
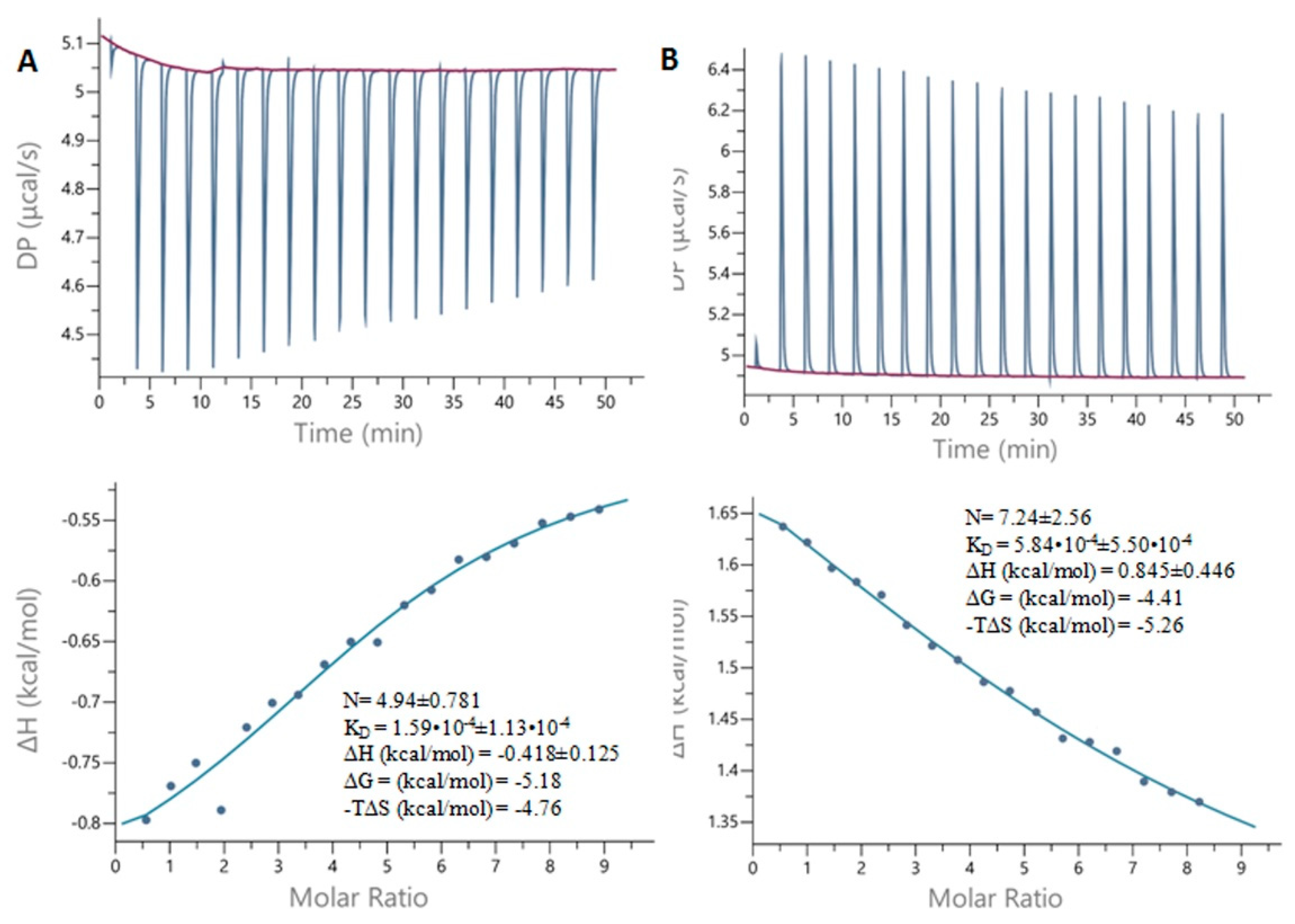
2.7. Isothermal Titration Calorimetry
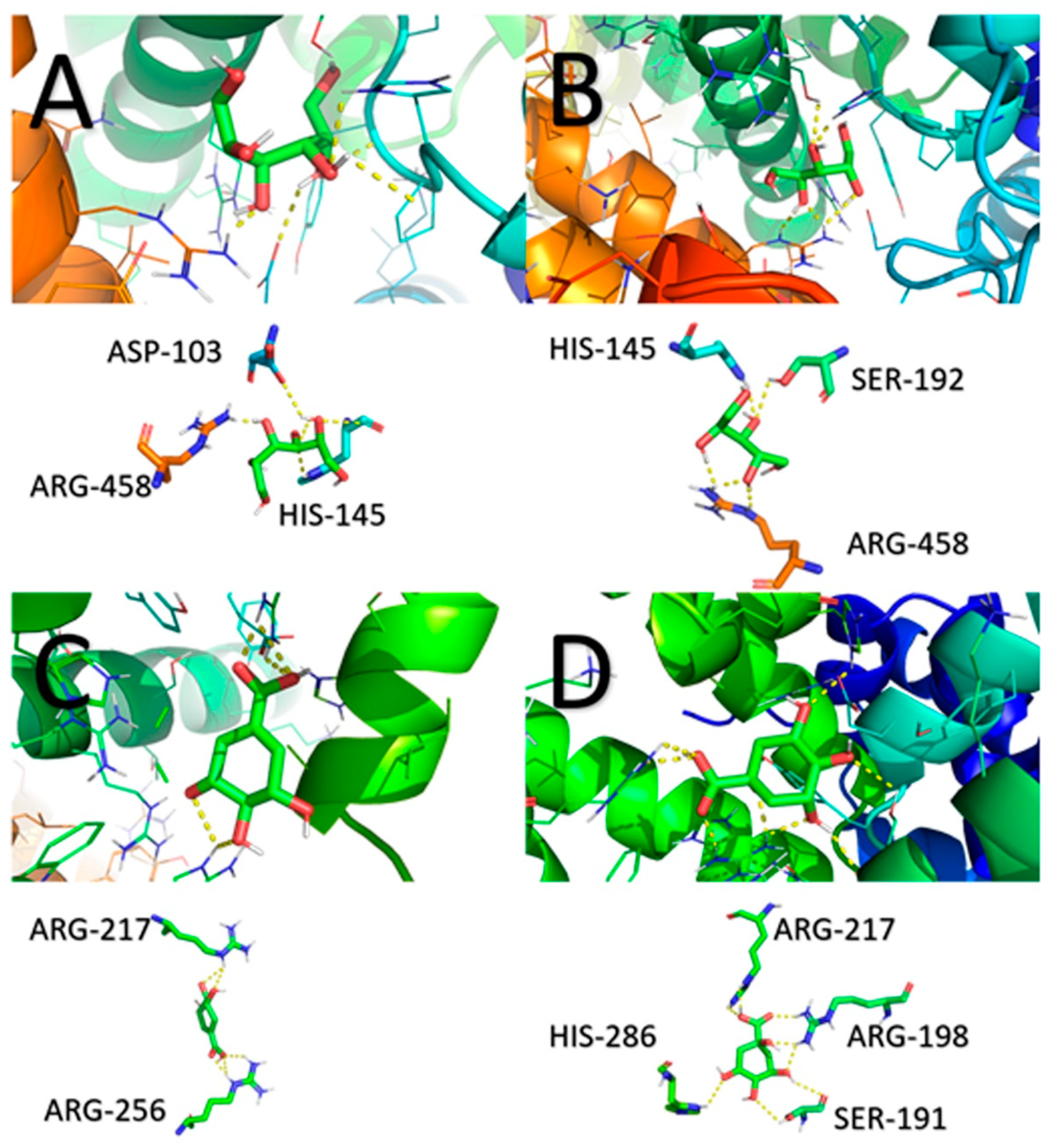
2.8. Molecular Docking
3. Materials and Methods
3.1. Materials
3.2. Preparation of BSA Solution
3.3. Preparation of Cyclitol Solutions
3.4. Sample Preparation for SDS-PAGE Separation
3.5. Kinetic Study of Cyclitol Binding to BSA
3.6. HPLC-MS/MS Analysis
3.6.1. Analysis of Protein–Cyclitol Interactions
3.6.2. Indication of Unbounded Cyclitols
3.7. Spectroscopic Analysis
3.7.1. UV-VIS Spectroscopy
3.7.2. Fluorescence Spectroscopy
3.7.3. Fourier Transform Infrared Spectroscopy (FTIR)
3.7.4. Raman Spectroscopy
3.8. Isothermal Titration Calorimetry
3.9. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Suod, H.; Ratiu, I.; Górecki, R.; Buszewski, B. Pressurized liquid extraction of cyclitols and sugars: Optimization of extraction parameters and selective separation. J. Sep. Sci. 2019, 42, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Antonowski, T.; Osowski, A.; Lahuta, L.; Górecki, R.; Rynkiewicz, A.; Wojtkiewicz, J. Health-Promoting Properties of Selected Cyclitols for Metabolic Syndrome and Diabetes. Nutrients 2019, 11, 2314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratiu, I.-A.; Al-Suod, H.; Ligor, M.; Ligor, T.; Railean-Plugaru, V.; Buszewski, B. Complex investigation of extraction techniques applied for cyclitols and sugars isolation from different species of Solidagogenus. Electrophoresis 2018, 39, 1966–1974. [Google Scholar] [CrossRef] [PubMed]
- Owczarczyk-Saczonek, A.; Lahuta, L.B.; Placek, W.; Górecki, R. The potential benefits of plant cyclitols in the treatment of psoriasis. Pol. Ann. Med. 2018, 25, 166–171. [Google Scholar] [CrossRef]
- Loescher, W.H. Physiology and metabolism of sugar alcohols in higher plants. Physiol. Plant. 1987, 70, 553–557. [Google Scholar] [CrossRef]
- Grembecka, M. Sugar alcohols—Their role in the modern world of sweeteners: A review. Eur. Food Res. Technol. 2015, 241, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chukwuma, C.; Islam, S. Sorbitol increases muscle glucose uptake ex vivo and inhibits intestinal glucose absorption ex vivo and in normal and type 2 diabetic rats. Appl. Physiol. Nutr. Metab. 2017, 42, 377–383. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.H.; Smith, D.C. Sugar Alcohols (Polyols) in Fungi and Green Plants. New Phytol. 1967, 66, 143–184. [Google Scholar] [CrossRef]
- de Valdéz, G.F.; de Giori, G.S.; Holgado, A.A.P.D.R.; Oliver, G. Protective Effect of Adonitol on Lactic Acid Bacteria Subjected to Freeze-Drying. Appl. Environ. Microbiol. 1983, 45, 302–304. [Google Scholar] [CrossRef] [Green Version]
- Cataldi, M.P.; Lu, P.; Blaeser, A.; Lu, Q.L. Ribitol restores functionally glycosylated α-dystroglycan and improves muscle function in dystrophic FKRP-mutant mice. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Escalante, A.A.; Carmona, S.B.; Quiroz, D.C.D.; Bolivar, F. Current perspectives on applications of shikimic and aminoshikimic acids in pharmaceutical chemistry. Res. Rep. Med. Chem. 2014, 4, 35–36. [Google Scholar] [CrossRef] [Green Version]
- Martínez, J.A.; Bolivar, F.; Escalante, A. Shikimic Acid Production in Escherichia coli: From Classical Metabolic Engineering Strategies to Omics Applied to Improve Its Production. Front. Bioeng. Biotechnol. 2015, 3, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, J.; Wu, Y.; Liu, X.; Zhong, K.; Huang, Y.; Gao, H. Antibacterial Activity of Shikimic Acid from Pine Needles of Cedrus deodara against Staphylococcus aureus through Damage to Cell Membrane. Int. J. Mol. Sci. 2015, 16, 27145–27155. [Google Scholar] [CrossRef] [Green Version]
- Heikkilä, E.; Hermant, A.; Thevenet, J.; Bermont, F.; Kulkarni, S.S.; Ratajczak, J.; Domingo, J.S.; Dioum, E.H.; Canto, C.; Barron, D.; et al. The plant product quinic acid activates Ca2+-dependent mitochondrial function and promotes insulin secretion from pancreatic beta cells. J. Cereb. Blood Flow Metab. 2019, 176, 3250–3263. [Google Scholar] [CrossRef]
- Pero, R.W.; Lund, H.; Leanderson, T. Antioxidant metabolism induced by quinic acid. increased urinary excretion of tryptophan and nicotinamide. Phytother. Res. 2008, 23, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.F.; Shi, L.P.; Ren, Y.D.; Liu, Q.F.; Liu, H.F.; Zhang, R.J.; Li, Z.; Zhu, F.H.; He, P.L.; Tang, W.; et al. Anti-Hepatitis B Virus Activity of Chlorogenic Acid, Quinic Acid and Caffeic Acid in Vivo and in Vitro. Antivir. Res. 2009, 83, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Naveenraj, S.; Anandan, S. Binding of serum albumins with bioactive substances—Nanoparticles to drugs. J. Photochem. Photobiol. C Photochem. Rev. 2013, 14, 53–71. [Google Scholar] [CrossRef]
- Squire, P.G.; Moser, P.; O’Konski, C.T. Hydrodynamic properties of bovine serum albumin monomer and dimer. Biochemistry 1968, 7, 4261–4272. [Google Scholar] [CrossRef]
- Yu, J.; Chen, Y.; Xiong, L.; Zhang, X.; Zheng, Y. Conductance Changes in Bovine Serum Albumin Caused by Drug-Binding Triggered Structural Transitions. Materials 2019, 12, 1022. [Google Scholar] [CrossRef] [Green Version]
- Lemli, B.; Derdák, D.; Laczay, P.; Kovács, D.; Kunsági-Máté, S. Noncovalent Interaction of Tilmicosin with Bovine Serum Albumin. Molecules 2018, 23, 1915. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulou, A.; Green, R.J.; Frazier, R.A. Interaction of Flavonoids with Bovine Serum Albumin: A Fluorescence Quenching Study. J. Agric. Food Chem. 2004, 53, 158–163. [Google Scholar] [CrossRef]
- Choi, J.-K.; Ho, J.; Curry, S.; Qin, D.; Bittman, R.; Hamilton, J. Interactions of very long-chain saturated fatty acids with serum albumin. J. Lipid Res. 2002, 43, 1000–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Das, T.K. Investigation of Binding of Bovine Serum Albumin with Metallic Nanoparticles. J. Chem. Pharm. Res. 2015, 7, 1203–1212. [Google Scholar]
- Guryanov, I.; Fiorucci, S.; Tennikova, T. Receptor-ligand interactions: Advanced biomedical applications. Mater. Sci. Eng. C 2016, 68, 890–903. [Google Scholar] [CrossRef]
- Tromelin, A.; Andriot, I.; Guichard, E. Protein-Flavour Interactions. In Flavour in Food; Voilley, A., Etievant, P.X., Eds.; Woodhead Publishing Series in Food Science: Cambridge, UK, 2006; pp. 172–207. [Google Scholar] [CrossRef]
- Gallina, A.M.; Bork, P.; Bordo, D. Structural analysis of protein-ligand interactions: The binding of endogenous compounds and of synthetic drugs. J. Mol. Recognit. 2014, 27, 65–72. [Google Scholar] [CrossRef]
- Bapli, A.; Chatterjee, A.; Gautam, R.K.; Jana, R.; Seth, D. Modulation of the Protein–Ligand Interaction in the Presence of Graphene Oxide: A Detailed Spectroscopic Study. Langmuir 2021, 37, 5034–5048. [Google Scholar] [CrossRef]
- Topală, T.; Bodoki, A.; Oprean, L.; Oprean, R. Bovine serum albumin interactions with metal complexes. Med. Pharm. Rep. 2014, 87, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Gekko, K.; Morikawa, T. Preferential Hydration of Bovine Serum Albumin in Polyhydric Alcohol-Water Mixtures1. J. Biochem. 1981, 90, 39–50. [Google Scholar] [CrossRef]
- Liu, Y.; Bolen, D.W. The Peptide Backbone Plays a Dominant Role in Protein Stabilization by Naturally Occurring Osmolytes. Biochemistry 1995, 34, 12884–12891. [Google Scholar] [CrossRef]
- González, M.; Murature, D.A.; Fidelio, G.D. Thermal Stability of Human Immunoglobulins with Sorbitol. Vox Sang. 1995, 68, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, P.; Tran, A.; Labuza, T.P. Effects of Polyols on the Stability of Whey Proteins in Intermediate-Moisture Food Model Systems. J. Agric. Food Chem. 2009, 57, 2339–2345. [Google Scholar] [CrossRef] [PubMed]
- Pomastowski, P.; Sprynskyy, M.; Buszewski, B. The study of zinc ions binding to casein. Colloids Surf. B Biointerfaces 2014, 120, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Buszewski, B.; Žuvela, P.; Król-Górniak, A.; Railean-Plugaru, V.; Rogowska, A.; Wong, M.W.; Yi, M.; Rodzik, A.; Sprynskyy, M.; Pomastowski, P. Interactions of zinc aqua complexes with ovalbumin at the forefront of the Zn2+/ZnO-OVO hybrid complex formation mechanism. Appl. Surf. Sci. 2020, 542, 148641. [Google Scholar] [CrossRef]
- Chanasattru, W.; Decker, E.; McClements, D.J. Impact of cosolvents (polyols) on globular protein functionality: Ultrasonic velocity, density, surface tension and solubility study. Food Hydrocoll. 2007, 22, 1475–1484. [Google Scholar] [CrossRef]
- Shimizua, S.; Smith, D.J. Preferential hydration and the exclusion of cosolvents from protein surfaces. J. Chem. Phys. 2004, 121, 1148–1154. [Google Scholar] [CrossRef]
- Rodzik, A.; Pomastowski, P.; Railean-Plugaru, V.; Sprynskyy, M.; Buszewski, B. The Study of Zinc Ions Binding to αS1-, β- and κ-Casein. Int. J. Mol. Sci. 2020, 21, 8096. [Google Scholar] [CrossRef]
- Pomastowski, P.; Sprynskyy, M.; Žuvela, P.; Rafińska, K.; Milanowski, M.; Liu, J.J.; Yi, M.; Buszewski, B. Silver-Lactoferrin Nanocomplexes as a Potent Antimicrobial Agent. J. Am. Chem. Soc. 2016, 138, 7899–7909. [Google Scholar] [CrossRef]
- Valderrama, C.; Gamisans, X.; Heras, X.D.L.; Farrán, A.; Cortina, J. Sorption kinetics of polycyclic aromatic hydrocarbons removal using granular activated carbon: Intraparticle diffusion coefficients. J. Hazard. Mater. 2008, 157, 386–396. [Google Scholar] [CrossRef]
- Hlavacek, W.S.; Posner, R.G.; Perelson, A.S. Steric Effects on Multivalent Ligand-Receptor Binding: Exclusion of Ligand Sites by Bound Cell Surface Receptors. Biophys. J. 1999, 76, 3031–3043. [Google Scholar] [CrossRef] [Green Version]
- Pryshchepa, O.; Sagandykova, G.N.; Pomastowski, P.; Railean-Plugaru, V.; Król, A.; Rogowska, A.; Rodzik, A.; Sprynskyy, M.; Buszewski, B. A New Approach for Spontaneous Silver Ions Immobilization onto Casein. Int. J. Mol. Sci. 2019, 20, 3864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gohlke, H.; Klebe, G. Approaches to the Description and Prediction of the Binding Affinity of Small-Molecule Ligands to Macromolecular Receptors. Angew. Chem. Int. Ed. 2002, 41, 2644–2676. [Google Scholar] [CrossRef]
- Zamora, I.; Oprea, T.; Cruciani, G.; Pastor, M.; Ungell, A.-L. Surface Descriptors for Protein−Ligand Affinity Prediction. J. Med. Chem. 2002, 46, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Q.; Zhang, W.; Jiang, Y.; Jiang, L.; Chen, X.; Liu, B. Study on the Interactions Between Caffeoylquinic Acids with Bovine Serum Albumin: Spectroscopy, Antioxidant Activity, LC-MSn, and Molecular Docking Approach. Front. Chem. 2019, 7, 840. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, R.F.; Schapira, M. A systematic analysis of atomic protein–ligand interactions in the PDB. MedChemComm 2017, 8, 1970–1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushik, J.; Bhat, R. Thermal Stability of Proteins in Aqueous Polyol Solutions: Role of the Surface Tension of Water in the Stabilizing Effect of Polyols. J. Phys. Chem. B 1998, 102, 7058–7066. [Google Scholar] [CrossRef]
- Buszewski, B.; Pomastowski, P. Influence of heterogeneity biocolloids surface om their electrophoretic separation. Wiadomości Chem. 2015, 69, 9–10. [Google Scholar]
- Ghisaidoobe, A.B.T.; Chung, S.J. Intrinsic Tryptophan Fluorescence in the Detection and Analysis of Proteins: A Focus on Förster Resonance Energy Transfer Techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Bortolotti, A.; Wong, Y.H.; Korsholm, S.S.; Bahring, N.H.B.; Bobone, S.; Tayyab, S.; van de Weert, M.; Stella, L. On the purported “backbone fluorescence” in protein three-dimensional fluorescence spectra. RSC Adv. 2016, 6, 112870–112876. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, S.; Ameen, F.; Rehman, S.U.; Sarwar, T.; Tabish, M. Studying the interaction of drug/ligand with serum albumin. J. Mol. Liq. 2021, 336, 116200. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohman, A.; Windarsih, A.; Lukitaningsih, E.; Rafi, M.; Betania, K.; Fadzillah, N.A. The use of FTIR and Raman spectroscopy in combination with chemometrics for analysis of biomolecules in biomedical fluids: A review. Biomed. Spectrosc. Imaging 2020, 8, 55–71. [Google Scholar] [CrossRef] [Green Version]
- Invernizzi, C.; Rovetta, T.; Licchelli, M.; Malagodi, M. Mid and Near-Infrared Reflection Spectral Database of Natural Organic Materials in the Cultural Heritage Field. Int. J. Anal. Chem. 2018, 2018, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Wang, J.; Yan, X.; Liu, R.; Sun, Y.; Li, X. Study of interaction between human serum albumin and three phenanthridine derivatives: Fluorescence spectroscopy and computational approach. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 145, 473–481. [Google Scholar] [CrossRef]
- Bouhekka, A.; Bürgi, T. In situ ATR-IR spectroscopy study of adsorbed protein: Visible light denaturation of bovine serum albumin on TiO2. Appl. Surf. Sci. 2012, 261, 369–374. [Google Scholar] [CrossRef]
- Alhazmi, H.A. FT-IR Spectroscopy for the Identification of Binding Sites and Measurements of the Binding Interactions of Important Metal Ions with Bovine Serum Albumin. Sci. Pharm. 2019, 87, 5. [Google Scholar] [CrossRef] [Green Version]
- Paschek, D.; Pühse, M.; Perez-Goicochea, A.; Gnanakaran, S.; García, A.E.; Winter, R.; Geiger, A. The Solvent-Dependent Shift of the Amide I Band of a Fully Solvated Peptide as a Local Probe for the Solvent Composition in the Peptide/Solvent Interface. ChemPhysChem 2008, 9, 2742–2750. [Google Scholar] [CrossRef]
- Barth, A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000, 74, 141–173. [Google Scholar] [CrossRef]
- Pinto, S.M.V.; Tasinato, N.; Barone, V.; Amadei, A.; Zanetti-Polzi, L.; Daidone, I. Modeling amino-acid side chain infrared spectra: The case of carboxylic residues. Phys. Chem. Chem. Phys. 2020, 22, 3008–3016. [Google Scholar] [CrossRef]
- Roselli, C.; Boussac, A.; Mattioli, T.A. Direct vibrational structure of protein metal-binding sites from near-infrared Yb3+ vibronic side band spectroscopy. Proc. Natl. Acad. Sci. USA 1994, 91, 12897–12901. [Google Scholar] [CrossRef] [Green Version]
- Sadat, A.; Joye, I.J. Peak Fitting Applied to Fourier Transform Infrared and Raman Spectroscopic Analysis of Proteins. Appl. Sci. 2020, 10, 5918. [Google Scholar] [CrossRef]
- Carpenter, J.F.; Crowe, J.H. An infrared spectroscopic study of the interactions of carbohydrates with dried proteins. Biochemistry 1989, 28, 3916–3922. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, J. Understanding the Amide-II Vibrations in β-Peptides. J. Phys. Chem. B 2015, 119, 14831–14839. [Google Scholar] [CrossRef] [PubMed]
- Žuvela, P.; Liu, J.J.; Yi, M.; Pomastowski, P.P.; Sagandykova, G.; Belka, M.; David, J.; Bączek, T.; Szafrański, K.; Żołnowska, B.; et al. Target-based drug discovery through inversion of quantitative structure-drug-property relationships and molecular simulation: CA IX-sulphonamide complexes. J. Enzym. Inhib. Med. Chem. 2018, 33, 1430–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stani, C.; Vaccari, L.; Mitri, E.; Birarda, G. FTIR investigation of the secondary structure of type I collagen: New insight into the amide III band. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 229, 1–7. [Google Scholar] [CrossRef]
- Pryshchepa, O.; Sagandykova, G.; Rudnicka, J.; Pomastowski, P.; Sprynskyy, M.; Buszewski, B. Synthesis and physicochemical characterization of zinc-lactoferrin complexes. J. Dairy Sci. 2022, 105, 1940–1958. [Google Scholar] [CrossRef]
- Qu, H.; Ling, Z.; Qi, X.; Xin, Y.; Liu, C.; Cao, H. A Remote Raman System and Its Applications for Planetary Material Studies. Sensors 2021, 21, 6973. [Google Scholar] [CrossRef]
- Trivedi, M.K.; Patil, S.; Shettigar, H.; Bairwa, K.; Jana, S. Spectroscopic Characterization of Biofield Treated Metronidazole and Tinidazole. Med. Chem. 2015, 5, 340–344. [Google Scholar] [CrossRef]
- Anguebes-Franseschi, F.; Abatal, M.; Pat, L.; Flores, A.; Quiroz, A.V.C.; Ramírez-Elias, M.A.; Pedro, L.S.; Tzuc, O.M.; Bassam, A. Raman Spectroscopy and Chemometric Modeling to Predict Physical-Chemical Honey Properties from Campeche, Mexico. Molecules 2019, 24, 4091. [Google Scholar] [CrossRef] [Green Version]
- Kuhar, N.; Sil, S.; Verma, T.; Umapathy, S. Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 2018, 8, 25888–25908. [Google Scholar] [CrossRef] [Green Version]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Kong, J.; Yu, S. Fourier Transform Infrared Spectroscopic Analysis of Protein Secondary Structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsztyńska-Janus, S.; Szymborska-Małek, K.; Gasior-Glogowska, M.; Walski, T.; Komorowska, M.; Witkiewicz, W.; Pezowicz, C.; Kobielarz, M.; Szotek, S. Spectroscopic Techniques in the Study of Human Tissues and Their Components. Part I: IR Spectroscopy. Acta Bioeng. Biomech. 2012, 14, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Singh, B.R. A Distinct Utility of the Amide III Infrared Band for Secondary Structure Estimation of Aqueous Protein Solutions Using Partial Least Squares Methods. Biochemistry 2004, 43, 2541–2549. [Google Scholar] [CrossRef]
- Reddy, S.B.; Nagaraja, M.; Kadalli, G.; Champa, B. Fourier Transform Infrared (FTIR) Spectroscopy of Soil Humic and Fulvic Acids Extracted from Paddy Land Use System. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 834–837. [Google Scholar] [CrossRef]
- Arsiccio, A.; Ganguly, P.; La Cortiglia, L.; Shea, J.-E.; Pisano, R. ADD Force Field for Sugars and Polyols: Predicting the Additivity of Protein–Osmolyte Interaction. J. Phys. Chem. B 2020, 124, 7779–7790. [Google Scholar] [CrossRef]
- Cloutier, T.; Sudrik, C.; Sathish, H.A.; Trout, B.L. Kirkwood–Buff-Derived Alcohol Parameters for Aqueous Carbohydrates and Their Application to Preferential Interaction Coefficient Calculations of Proteins. J. Phys. Chem. B 2018, 122, 9350–9360. [Google Scholar] [CrossRef]
- Di Muzio, E.; Toti, D.; Polticelli, F. DockingApp: A user friendly interface for facilitated docking simulations with AutoDock Vina. J. Comput. Mol. Des. 2017, 31, 213–218. [Google Scholar] [CrossRef]
- Tanchuk, V.Y.; Tanin, V.O.; Vovk, A.I.; Poda, G. A New, Improved Hybrid Scoring Function for Molecular Docking and Scoring Based on AutoDock and AutoDock Vina. Chem. Biol. Drug Des. 2015, 87, 618–625. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into Protein–Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Dash, S.; Murthy, P.N.; Nath, L.; Chowdhury, P. Kinetic modeling on drug release from controlled drug delivery systems. Acta Pol. Pharm. 2010, 67, 217–223. [Google Scholar] [PubMed]
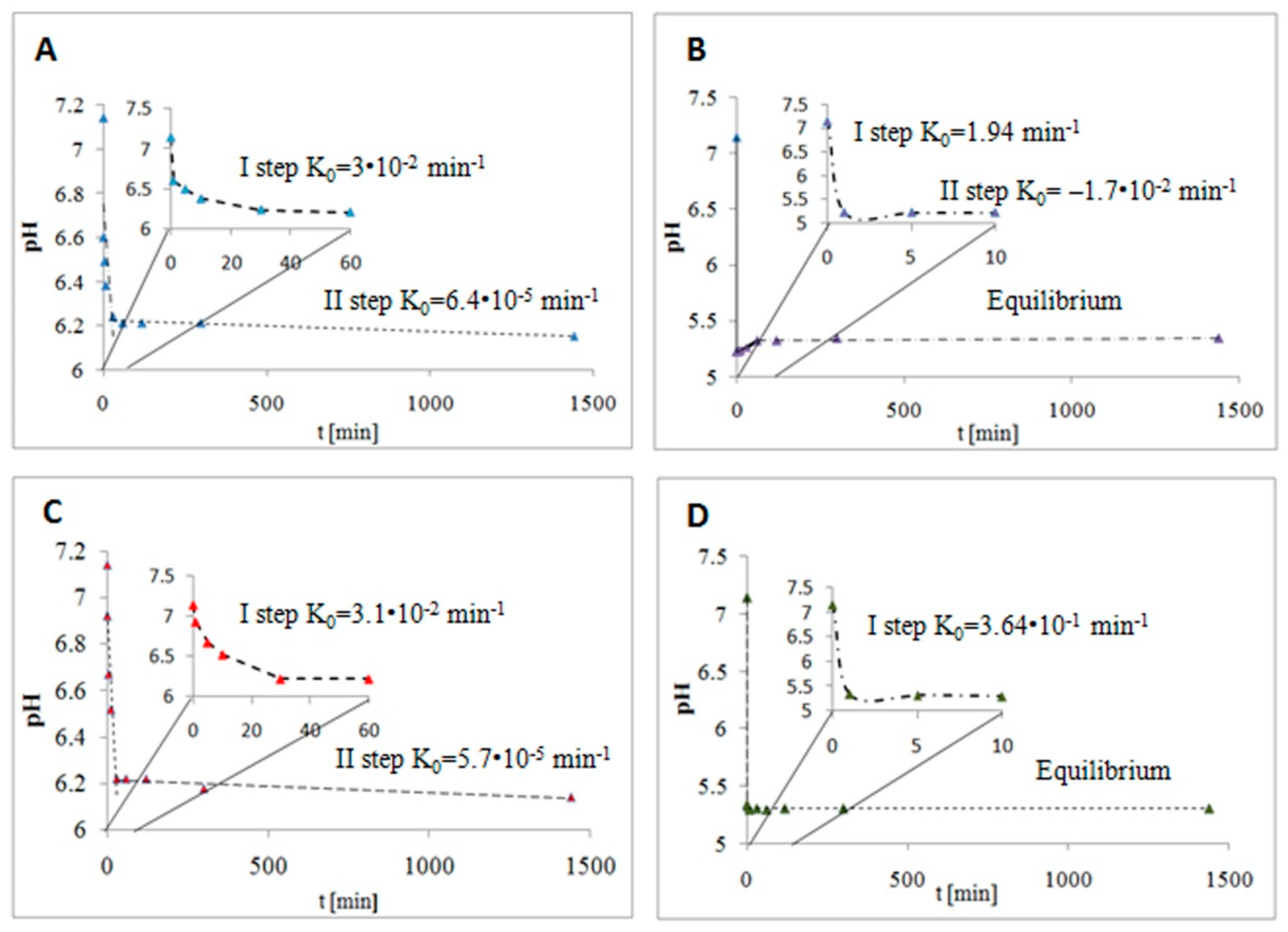
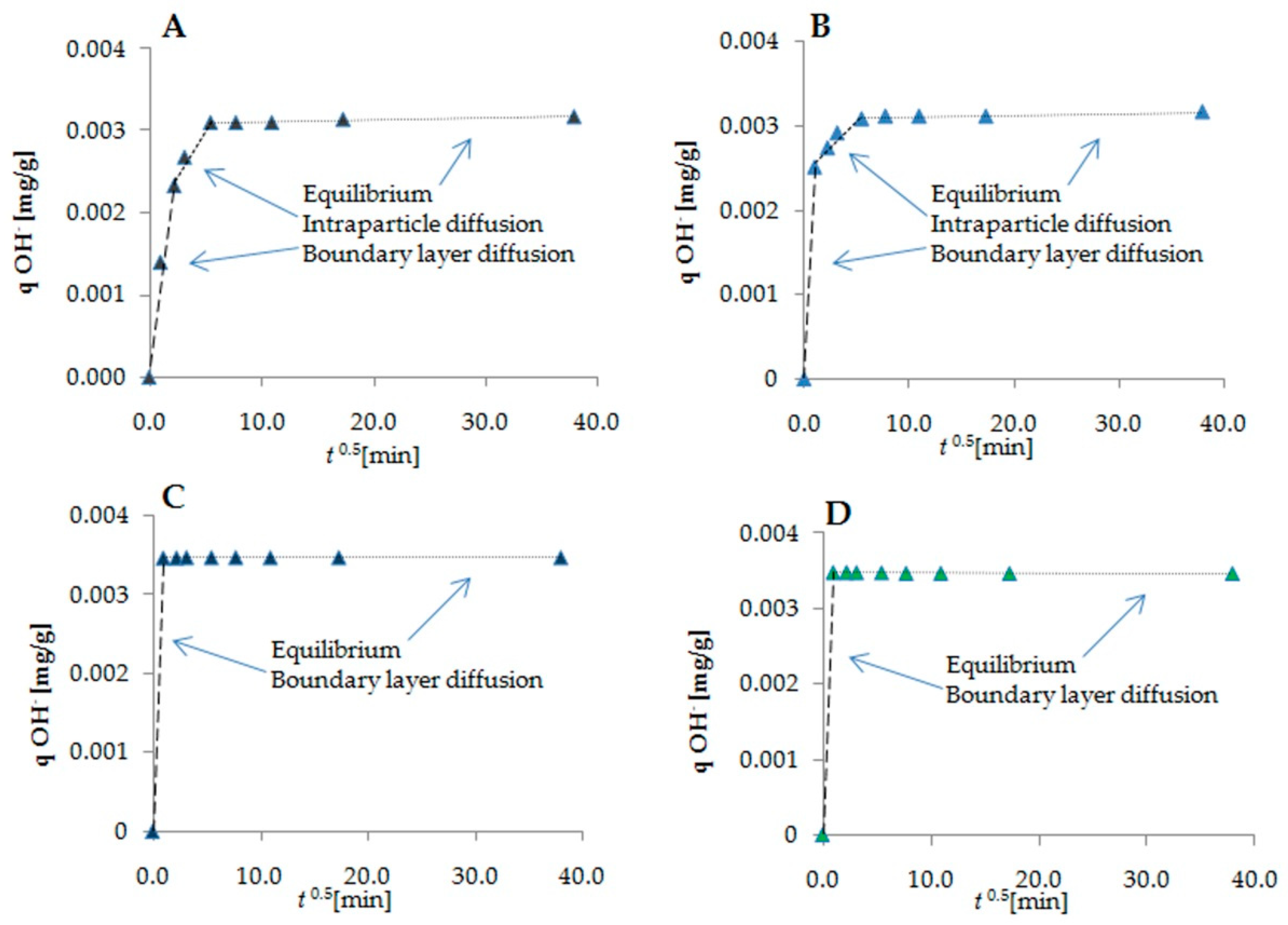
| Cyclitol | Zero-Order Kinetic Model | Intraparticle Diffusion Model | G (kJ/mol)/(kcal/mol) | ||
|---|---|---|---|---|---|
| D-sorbitol | Step I | 0.031 | A | 0.00199 | −21.74/−5.20 |
| k0 (min−1) | (mg/g) | ||||
| Step II | 0.000057 | Kip | 0.0000477 | ||
| k0 (min−1) | ((mg/g)/t0.5) | ||||
| Adonitol | Step I | 0.030 | A | 0.00230 | −21.69/−5.18 |
| k0 (min−1) | (mg/g) | ||||
| Step II | 0.000064 | Kip | 0.0000352 | ||
| k0 (min−1) | ((mg/g)/t0.5) | ||||
| Shikimic acid | Step I | 0.364 | A | 0.00281 | −26.37/−6.30 |
| k0 (min−1) | (mg/g) | ||||
| Kip | 0.0000289 | ||||
| ((mg/g)/t0.5) | |||||
| D-(–)-quinic acid | Step I | 1.94 | A | 0.00282 | −26.16/−6.25 |
| k0 (min−1) | (mg/g) | ||||
| Step II | −0.017 | Kip | 0.0000285 | ||
| k0 (min−1) | ((mg/g)/t0.5) | ||||
| Composition of Mobile Phase | tR | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Adonitol m/z (+) = 153 | D-Sorbitol m/z (–) = 181 | Shikimic Acid m/z (–) = 173 | D-(–)-Quinic Acid m/z (–) = 191 | ||||||
| [M − H]+ | [M − Na]+ | [2M − Na]+ | [M]− | [2M]− | [M]− | [2M]− | [M]− | [2M]− | |
| Water | 2.645 | 2.641 | - | 1.516 | - | - | - | - | - |
| 2.638 | |||||||||
| Water + 0.1% v/v FA | 2.626 | 2.626 | - | 1.550 | 1.550 | 5.099 | - | 10.273 | - |
| 2.621 | |||||||||
| Water + 0.5% v/v IP | 2.792 | 2.788 | 2.786 | 1.726 | 2.821 | - | - | - | 1.769 |
| 2.822 | |||||||||
| Water + 1% v/v IP | 2.903 | 2.900 | - | 1.672 | 2.888 | - | - | - | 1.721 |
| 2.887 | |||||||||
| Water + 2% v/v IP | 2.989 | 2.989 | - | 1.656 | 2.983 | - | - | - | 1.721 |
| 2.983 | |||||||||
| Water + 0.1% v/v FA + 0.5 % v/v IP | 2.630 | 2.630 | - | 1.490 | - | 5.064 | - | 10.269 | - |
| 2.623 | |||||||||
| Water + 0.1% v/v FA + 1% v/v IP | 2.641 | 2.638 | 2.632 | 1.559 | 1.561 | 5.060 | - | 10.297 | - |
| 2.631 | |||||||||
| Water + 0.1% v/v FA + 2% v/v IP | 2.635 | 2.635 | 2.635 | 1.551 | 1.554 | 5.057 | - | 10.344 | - |
| 2.644 | |||||||||
| Analyzed Cyclitol | m/z | Protein–Cyclitol Molar Ratio | Bounded Cyclitol with Protein ± SD | Equation of the Calibration Curve y = ax + b (R2) | Number of Bounded Ligand Molecules ± SD |
|---|---|---|---|---|---|
| D-sorbitol | [M]− = 181 | 1:2 | 49.92 ± 0.29 | y = 4.00 × 1011 + 506,935 (0.9985) | 0.99 ± 0.01 |
| 1:5 | 17.75 ± 1.02 | 0.89 ± 0.01 | |||
| 1:7 | 23.92 ± 1.13 | 1.67 ± 0.08 | |||
| 1:10 | 20.89 ± 1.25 | 2.09 ± 0.13 | |||
| Shikimic acid | [M]− = 173 | 1:2 | 92.56 ± 1.49 | y = 1.29 × 1011 + 85,318 (0.9982) | 1.85 ± 0.03 |
| 1:5 | 2.75 ± 0.09 | 54.98 ± 1.77 | |||
| 1:7 | 3.20 ± 0.02 | 45.70 ± 0.31 | |||
| D-(–)-quinic acid | [M]− = 191 | 1:2 | 3.95 ± 0.44 | y = 5.00 × 1011 + 433,847 (0.9960) | 0.08 ± 0.01 |
| 1:7 | 32.55 ± 1.31 | 2.28 ± 0.09 | |||
| 1:10 | 39.27 ± 0.94 | 3.93 ± 0.09 |
| Protein Receptor | Ligand | Amino Acid Residues | Number of Bonds | Bond Length (A°) | Calculated Binding Energy of the First Ligand Molecule to BSA (kcal/mol) |
|---|---|---|---|---|---|
| BSA | D-sorbitol | Arg458 | 2 | 3.2/1.9 | |
| His145 | 1 | 2.8 | −5.5 | ||
| Asp103 | 1 | 2.4 | |||
| BSA | Adonitol | Arg458 | 3 | 1.2/2.1/2.5 | |
| Ser192 | 1 | 2.4 | −5.0 | ||
| His145 | 1 | 2.0 | |||
| BSA | Shikimic acid | Arg217 | 2 | 2.6/2.8 | −5.7 |
| Arg256 | 3 | 2.1/2.5/2.3 | |||
| BSA | D-(–)-quinic acid | Arg217 | 2 | 2.3/2.4 | |
| Arg198 | 3 | 2.1/2.1/2.3 | −6.0 | ||
| Ser191 | 2 | 2.9/2.6 | |||
| His286 | 2 | 3.3/2.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyrda-Terniuk, T.; Sugajski, M.; Pryshchepa, O.; Śliwiak, J.; Buszewska-Forajta, M.; Pomastowski, P.; Buszewski, B. The Study of Protein–Cyclitol Interactions. Int. J. Mol. Sci. 2022, 23, 2940. https://doi.org/10.3390/ijms23062940
Dyrda-Terniuk T, Sugajski M, Pryshchepa O, Śliwiak J, Buszewska-Forajta M, Pomastowski P, Buszewski B. The Study of Protein–Cyclitol Interactions. International Journal of Molecular Sciences. 2022; 23(6):2940. https://doi.org/10.3390/ijms23062940
Chicago/Turabian StyleDyrda-Terniuk, Tetiana, Mateusz Sugajski, Oleksandra Pryshchepa, Joanna Śliwiak, Magdalena Buszewska-Forajta, Paweł Pomastowski, and Bogusław Buszewski. 2022. "The Study of Protein–Cyclitol Interactions" International Journal of Molecular Sciences 23, no. 6: 2940. https://doi.org/10.3390/ijms23062940
APA StyleDyrda-Terniuk, T., Sugajski, M., Pryshchepa, O., Śliwiak, J., Buszewska-Forajta, M., Pomastowski, P., & Buszewski, B. (2022). The Study of Protein–Cyclitol Interactions. International Journal of Molecular Sciences, 23(6), 2940. https://doi.org/10.3390/ijms23062940









