Near-Infrared Markers based on Bacterial Phytochromes with Phycocyanobilin as a Chromophore
Abstract
1. Introduction
2. Results
2.1. The Interaction of iRFP713 and Its Mutant Variants with Phycocyanobilin Cofactor (PCB)
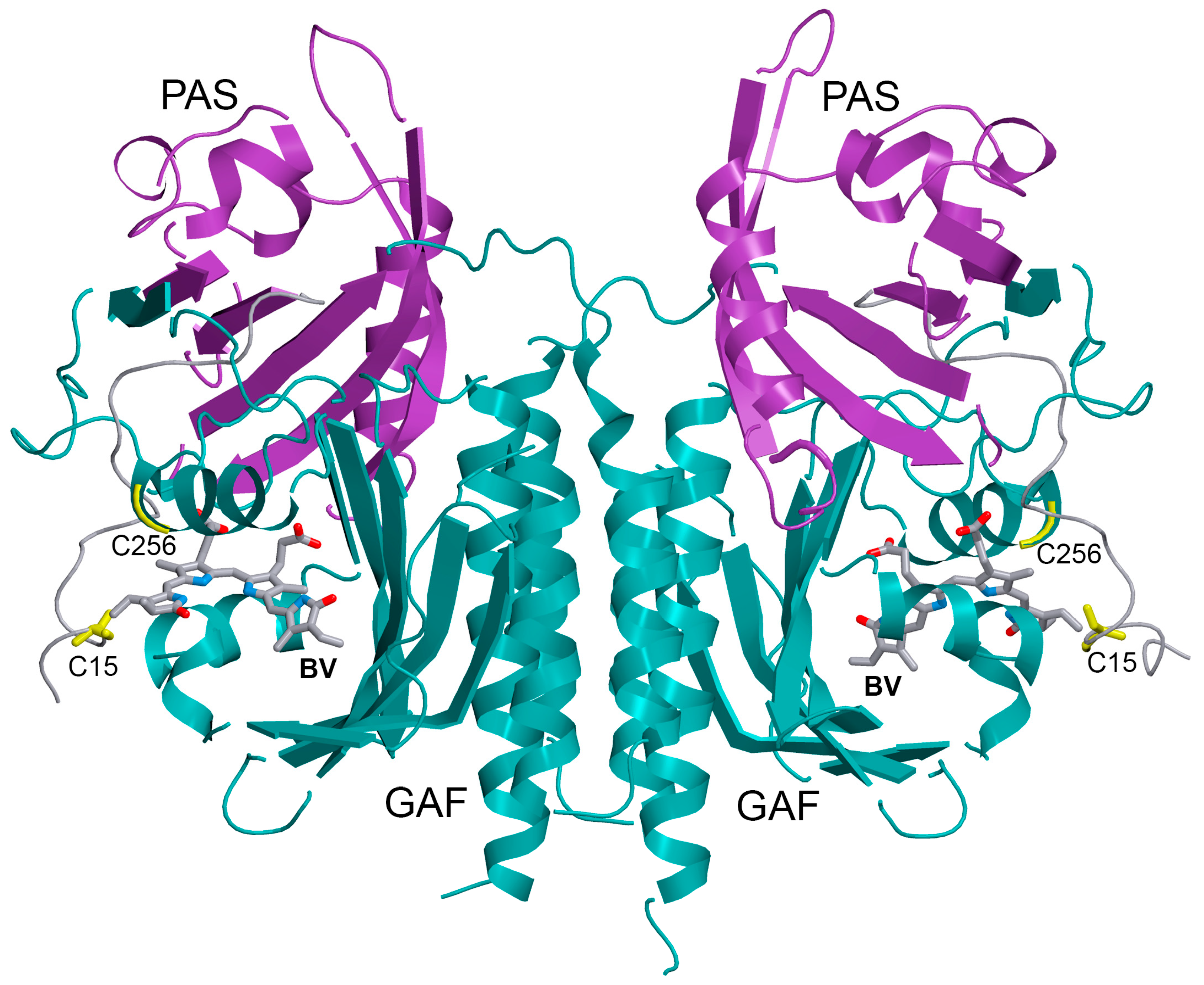
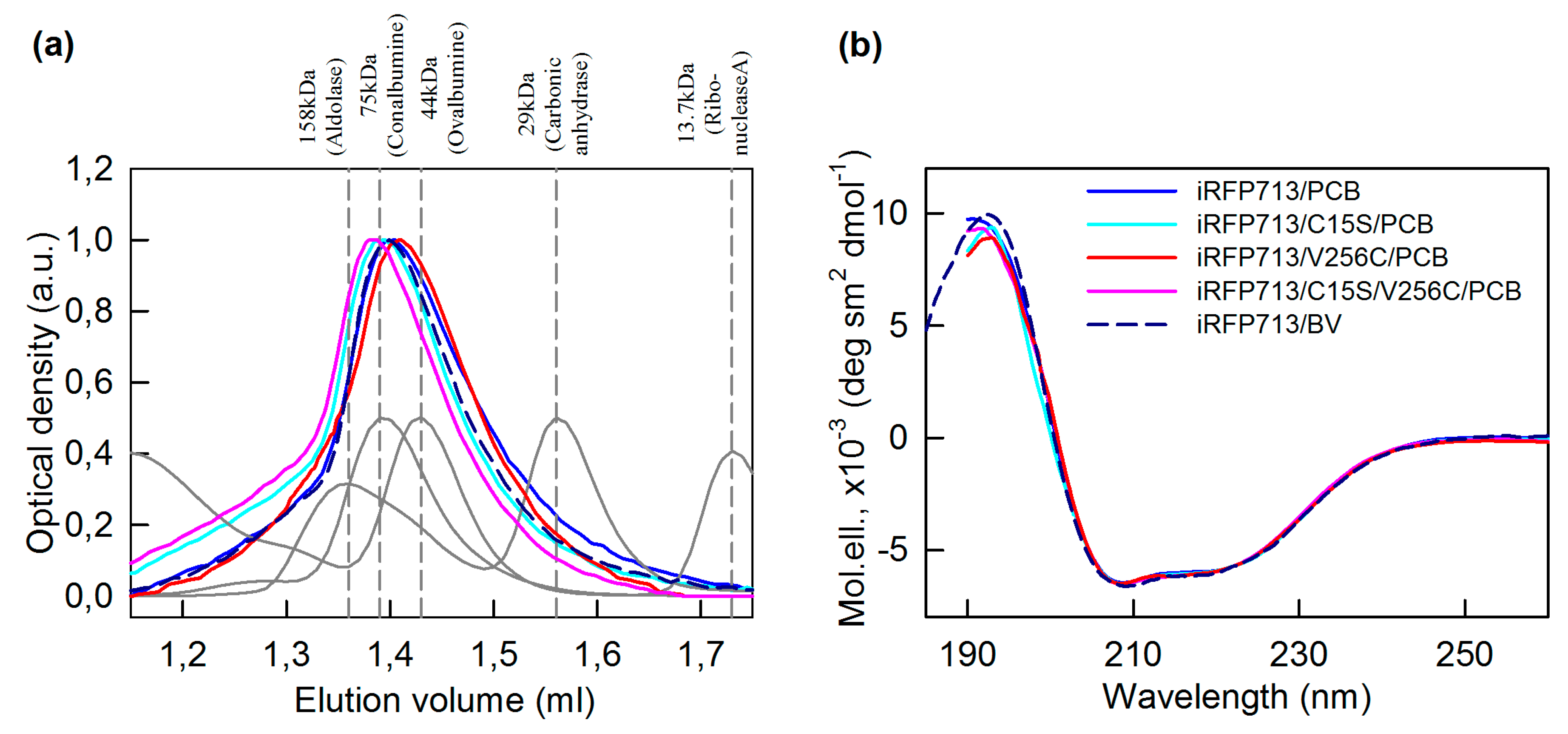
2.2. The Structure of iRFP713 and Its Mutant Variants Assembled with PCB
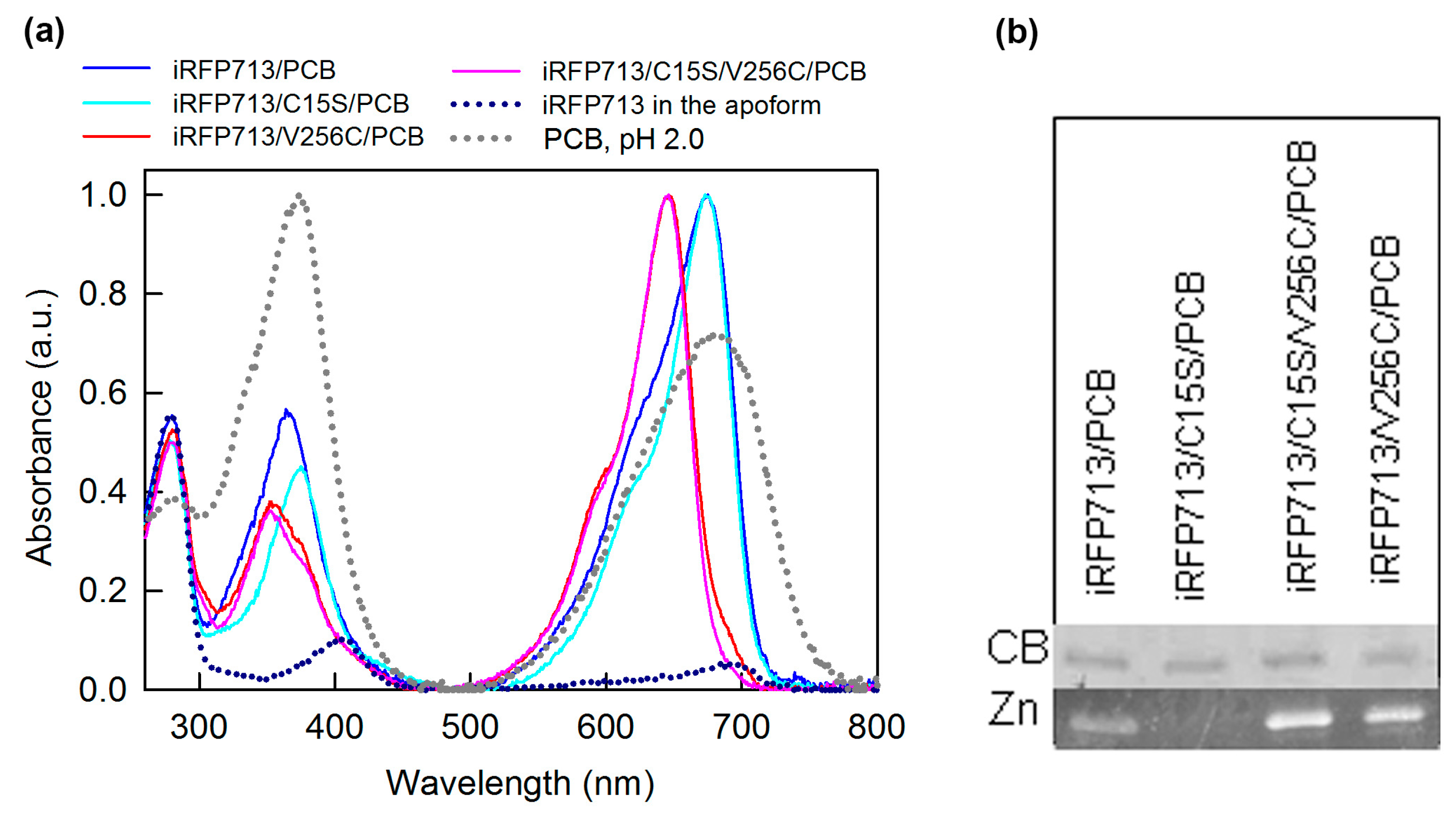
2.3. Spectral Properties of iRFP713 and Its Mutant Variants Assembled with PCB
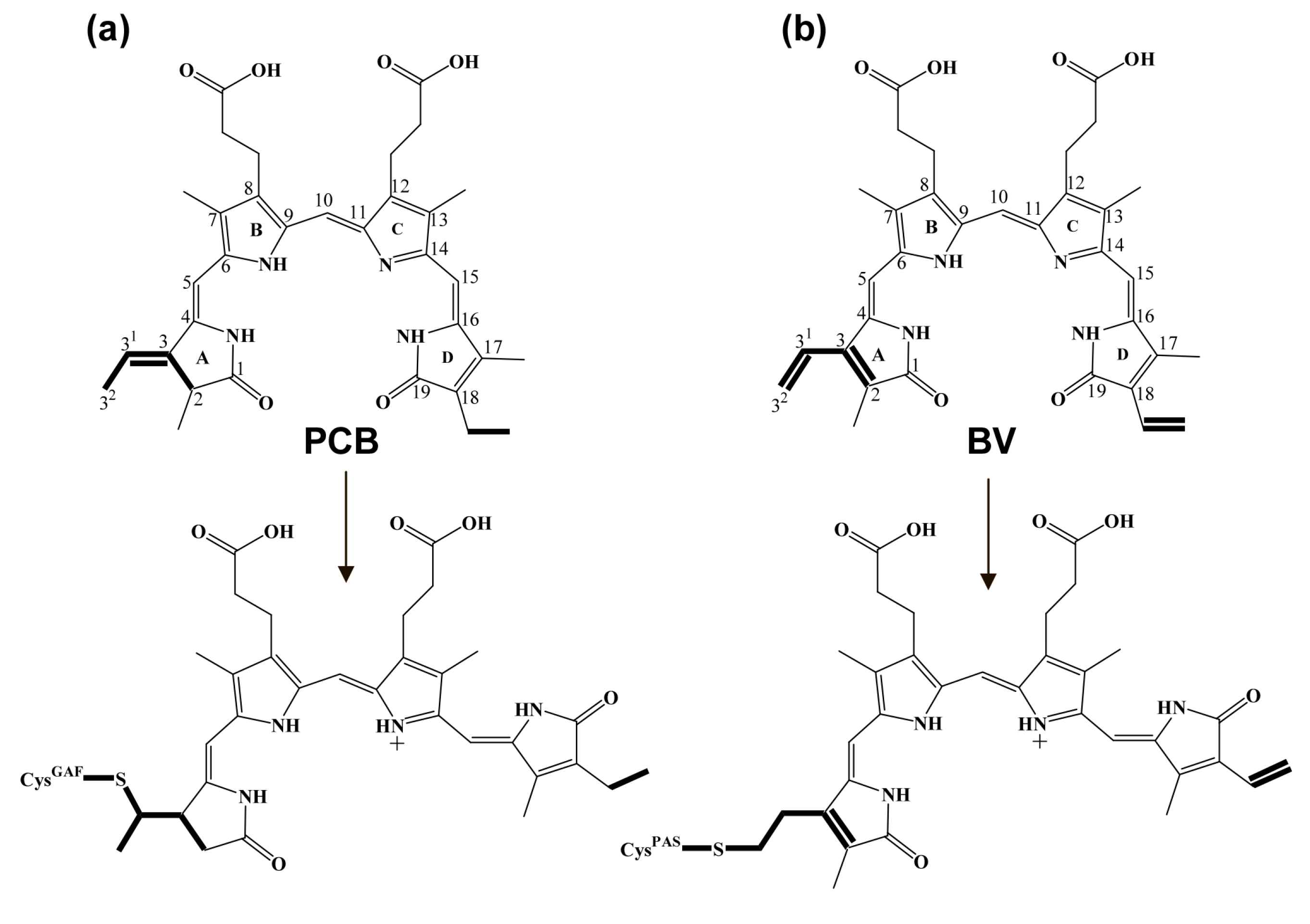
2.4. A Configuration of PCB Incorporated into iRFP713 and Its Mutant Variants
2.5. Competitive Binding of PCB and BV with iRFP713 and Its Variants
3. Discussion
4. Materials and Methods
4.1. Protein Expression and Purification
4.2. Spectral and Biochemical Characterization of Proteins
4.3. Analysis of the Chromophore Geometry
4.4. Gel Filtration Experiments
4.5. Circular Dichroism Measurements
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BDFP | “Big Dipper constellation” Fluorescent Protein |
| BV | biliverdin IXα |
| GAF | cGMP phosphodiesterase/Adenyl cyclase/FhlA domain |
| GFP | Green Fluorescent Protein |
| PAS | Per/Arnt/Sim domain |
| PCB | phycocyanobilin |
| BphP | bacterial phytochrome |
| NIR FP | near infrared fluorescent protein |
| CBCR | cyanobacteriophytochrome |
References
- Mishin, A.S.; Belousov, V.V.; Solntsev, K.M.; Lukyanov, K.A. Novel uses of fluorescent proteins. Curr. Opin. Chem. Biol. 2015, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Stepanenko, O.V.; Shcherbakova, D.M.; Kuznetsova, I.M.; Turoverov, K.K.; Verkhusha, V.V. Modern fluorescent proteins: From chromophore formation to novel intracellular applications. BioTechniques 2011, 51, 313–314, 316, 318 passim. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Karasev, M.M.; Stepanenko, O.V.; Rumyantsev, K.A.; Turoverov, K.K.; Verkhusha, V.V. Near-Infrared Fluorescent Proteins and Their Applications. Biochem. Biokhimiia 2019, 84, S32–S50. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Verkhusha, V.V. Near-Infrared Fluorescent Proteins: Multiplexing and Optogenetics across Scales. Trends Biotechnol. 2018, 36, 1230–1243. [Google Scholar] [CrossRef]
- Kapitulnik, J.; Maines, M.D. The role of bile pigments in health and disease: Effects on cell signaling, cytotoxicity, and cytoprotection. Front. Pharmacol. 2012, 3, 136. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Satyshur, K.A.; Anstrom, D.M.; Forest, K.T. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 2012, 287, 7000–7009. [Google Scholar] [CrossRef]
- Lehtivuori, H.; Bhattacharya, S.; Angenent-Mari, N.M.; Satyshur, K.A.; Forest, K.T. Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor. Front. Mol. Biosci. 2015, 2, 65. [Google Scholar] [CrossRef]
- Song, C.; Psakis, G.; Lang, C.; Mailliet, J.; Gartner, W.; Hughes, J.; Matysik, J. Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. USA 2011, 108, 3842–3847. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Auldridge, M.E.; Lehtivuori, H.; Ihalainen, J.A.; Forest, K.T. Origins of fluorescence in evolved bacteriophytochromes. J. Biol. Chem. 2014, 289, 32144–32152. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Baloban, M.; Bublikov, G.S.; Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Kuznetsova, I.M.; Verkhusha, V.V. Allosteric effects of chromophore interaction with dimeric near-infrared fluorescent proteins engineered from bacterial phytochromes. Sci. Rep. 2016, 6, 18750. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Shcherbakova, D.M.; Verkhusha, V.V.; Turoverov, K.K. Interaction of Biliverdin Chromophore with Near-Infrared Fluorescent Protein BphP1-FP Engineered from Bacterial Phytochrome. Int. J. Mol. Sci. 2017, 18, 1009. [Google Scholar] [CrossRef] [PubMed]
- Toh, K.C.; Stojkovic, E.A.; van Stokkum, I.H.; Moffat, K.; Kennis, J.T. Proton-transfer and hydrogen-bond interactions determine fluorescence quantum yield and photochemical efficiency of bacteriophytochrome. Proc. Natl. Acad. Sci. USA 2010, 107, 9170–9175. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Cox Cammer, N.; Huisman, T.M.; Verkhusha, V.V.; Hodgson, L. Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat. Chem. Biol. 2018, 14, 591–600. [Google Scholar] [CrossRef]
- Rodriguez, E.A.; Tran, G.N.; Gross, L.A.; Crisp, J.L.; Shu, X.; Lin, J.Y.; Tsien, R.Y. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods 2016, 13, 763–769. [Google Scholar] [CrossRef]
- Ding, W.L.; Miao, D.; Hou, Y.N.; Jiang, S.P.; Zhao, B.Q.; Zhou, M.; Scheer, H.; Zhao, K.H. Small monomeric and highly stable near-infrared fluorescent markers derived from the thermophilic phycobiliprotein, ApcF2. Biochim. Et Biophys. Acta. Mol. Cell Res. 2017, 1864, 1877–1886. [Google Scholar] [CrossRef]
- Hou, Y.N.; Ding, W.L.; Jiang, S.P.; Miao, D.; Tan, Z.Z.; Hu, J.L.; Scheer, H.; Zhao, K.H. Bright near-infrared fluorescence bio-labeling with a biliprotein triad. Biochim. Et Biophys. Acta. Mol. Cell Res. 2019, 1866, 277–284. [Google Scholar] [CrossRef]
- Kunkel, T.; Tomizawa, K.; Kern, R.; Furuya, M.; Chua, N.H.; Schafer, E. In vitro formation of a photoreversible adduct of phycocyanobilin and tobacco apophytochrome B. Eur. J. Biochem. 1993, 215, 587–594. [Google Scholar] [CrossRef]
- Beyer, H.M.; Juillot, S.; Herbst, K.; Samodelov, S.L.; Muller, K.; Schamel, W.W.; Romer, W.; Schafer, E.; Nagy, F.; Strahle, U.; et al. Red Light-Regulated Reversible Nuclear Localization of Proteins in Mammalian Cells and Zebrafish. ACS Synth. Biol. 2015, 4, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Buckley, C.E.; Moore, R.E.; Reade, A.; Goldberg, A.R.; Weiner, O.D.; Clarke, J.D.W. Reversible Optogenetic Control of Subcellular Protein Localization in a Live Vertebrate Embryo. Dev. Cell 2016, 36, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Muller, K.; Engesser, R.; Metzger, S.; Schulz, S.; Kampf, M.M.; Busacker, M.; Steinberg, T.; Tomakidi, P.; Ehrbar, M.; Nagy, F.; et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013, 41, e77. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jost, A.P.; Weiner, O.D.; Tang, C. A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol. Biol. Cell 2013, 24, 2419–2430. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhao, B.Q.; Miao, D.; Ding, W.L.; Zhou, M.; Scheer, H.; Zhao, K.H. A Simple Preparation Method for Phytochromobilin. Photochem. Photobiol. 2017, 93, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Oliinyk, O.S.; Shemetov, A.A.; Pletnev, S.; Shcherbakova, D.M.; Verkhusha, V.V. Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat. Commun. 2019, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Martin, S.S.; Lagarias, J.C. Identification of Cyanobacteriochromes Detecting Far-Red Light. Biochemistry 2016, 55, 3907–3919. [Google Scholar] [CrossRef]
- Filonov, G.S.; Piatkevich, K.D.; Ting, L.M.; Zhang, J.; Kim, K.; Verkhusha, V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011, 29, 757–761. [Google Scholar] [CrossRef]
- Rockwell, N.C.; Lagarias, J.C. A brief history of phytochromes. Chemphyschem 2010, 11, 1172–1180. [Google Scholar] [CrossRef]
- Bellini, D.; Papiz, M.Z. Dimerization properties of the RpBphP2 chromophore-binding domain crystallized by homologue-directed mutagenesis. Acta Cryst. D Biol Cryst. 2012, 68, 1058–1066. [Google Scholar] [CrossRef]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An introductory tutorial. Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef] [PubMed]
- Merritt, E.A.; Bacon, D.J. Raster3D: Photorealistic molecular graphics. Methods Enzym. 1997, 277, 505–524. [Google Scholar]
- Mukougawa, K.; Kanamoto, H.; Kobayashi, T.; Yokota, A.; Kohchi, T. Metabolic engineering to produce phytochromes with phytochromobilin, phycocyanobilin, or phycoerythrobilin chromophore in Escherichia coli. FEBS Lett 2006, 580, 1333–1338. [Google Scholar] [CrossRef]
- Lamparter, T.; Mittmann, F.; Gartner, W.; Borner, T.; Hartmann, E.; Hughes, J. Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proc. Natl. Acad. Sci. USA 1997, 94, 11792–11797. [Google Scholar] [CrossRef] [PubMed]
- Park, C.M.; Shim, J.Y.; Yang, S.S.; Kang, J.G.; Kim, J.I.; Luka, Z.; Song, P.S. Chromophore-apoprotein interactions in Synechocystis sp. PCC6803 phytochrome Cph1. Biochemistry 2000, 39, 6349–6356. [Google Scholar] [CrossRef]
- Wagner, J.R.; Zhang, J.; von Stetten, D.; Gunther, M.; Murgida, D.H.; Mroginski, M.A.; Walker, J.M.; Forest, K.T.; Hildebrandt, P.; Vierstra, R.D. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 2008, 283, 12212–12226. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Stepanenko, O.V.; Bublikov, G.S.; Kuznetsova, I.M.; Verkhusha, V.V.; Turoverov, K.K. Stabilization of structure in near-infrared fluorescent proteins by binding of biliverdin chromophore. J. Mol. Struct. 2017, 1140, 22–31. [Google Scholar] [CrossRef]
- Provencher, S.W.; Glockner, J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981, 20, 33–37. [Google Scholar] [CrossRef]
- Hahn, J.; Strauss, H.M.; Landgraf, F.T.; Gimenez, H.F.; Lochnit, G.; Schmieder, P.; Hughes, J. Probing protein-chromophore interactions in Cph1 phytochrome by mutagenesis. FEBS J. 2006, 273, 1415–1429. [Google Scholar] [CrossRef]
- Burgie, E.S.; Bussell, A.N.; Lye, S.H.; Wang, T.; Hu, W.; McLoughlin, K.E.; Weber, E.L.; Li, H.; Vierstra, R.D. Photosensing and Thermosensing by Phytochrome B Require Both Proximal and Distal Allosteric Features within the Dimeric Photoreceptor. Sci. Rep. 2017, 7, 13648. [Google Scholar] [CrossRef]
- Lamparter, T.; Michael, N.; Mittmann, F.; Esteban, B. Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc. Natl. Acad. Sci. USA 2002, 99, 11628–11633. [Google Scholar] [CrossRef] [PubMed]
- Quest, B.; Hubschmann, T.; Sharda, S.; Tandeau de Marsac, N.; Gartner, W. Homologous expression of a bacterial phytochrome. The cyanobacterium Fremyella diplosiphon incorporates biliverdin as a genuine, functional chromophore. FEBS J. 2007, 274, 2088–2098. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Zhang, J.; Brunzelle, J.S.; Vierstra, R.D.; Forest, K.T. High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 2007, 282, 12298–12309. [Google Scholar] [CrossRef] [PubMed]
- Knipp, B.; Müller, M.; Metzler-Nolte, N.; Balaban, T.S.; Braslavsky, S.E.; Schaffner, K. NMR Verification of Helical Conformations of Phycocyanobilin in Organic Solvents. Helv. Chim. Acta 1998, 81, 881–888. [Google Scholar] [CrossRef]
- Borucki, B.; Otto, H.; Rottwinkel, G.; Hughes, J.; Heyn, M.P.; Lamparter, T. Mechanism of Cph1 phytochrome assembly from stopped-flow kinetics and circular dichroism. Biochemistry 2003, 42, 13684–13697. [Google Scholar] [CrossRef]
- Lamparter, T.; Michael, N.; Caspani, O.; Miyata, T.; Shirai, K.; Inomata, K. Biliverdin binds covalently to agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 2003, 278, 33786–33792. [Google Scholar] [CrossRef]
- Khrenova, M.G.; Kulakova, A.M.; Nemukhin, A.V. Competition between two cysteines in covalent binding of biliverdin to phytochrome domains. Org. Biomol. Chem. 2018, 16, 7518–7529. [Google Scholar] [CrossRef]
- Loughlin, P.C.; Duxbury, Z.; Mugerwa, T.T.; Smith, P.M.; Willows, R.D.; Chen, M. Spectral properties of bacteriophytochrome AM1_5894 in the chlorophyll d-containing cyanobacterium Acaryochloris marina. Sci. Rep. 2016, 6, 27547. [Google Scholar] [CrossRef]
- Lehtivuori, H.; Rissanen, I.; Takala, H.; Bamford, J.; Tkachenko, N.V.; Ihalainen, J.A. Fluorescence properties of the chromophore-binding domain of bacteriophytochrome from Deinococcus radiodurans. J. Phys. Chem. B 2013, 117, 11049–11057. [Google Scholar] [CrossRef]
- Toh, K.C.; Stojkovic, E.A.; Rupenyan, A.B.; van Stokkum, I.H.; Salumbides, M.; Groot, M.L.; Moffat, K.; Kennis, J.T. Primary reactions of bacteriophytochrome observed with ultrafast mid-infrared spectroscopy. J. Phys. Chem. A 2011, 115, 3778–3786. [Google Scholar] [CrossRef]
- Favre, H.A.; Powell, W.H. Nomenclature of Organic Chemistry: IUPAC Recommendations and Preferred Names 2013; Royal Society of Chemistry: London, UK, 2014; p. 1568. [Google Scholar]
- Baloban, M.; Shcherbakova, D.M.; Pletnev, S.; Pletnev, V.Z.; Lagarias, J.C.; Verkhusha, V.V. Designing brighter near-infrared fluorescent proteins: Insights from structural and biochemical studies. Chem. Sci. 2017, 8, 4546–4557. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Ding, W.L.; Hoppner, A.; Zhao, C.; Zhang, L.; Hontani, Y.; Kennis, J.T.; Gartner, W.; Scheer, H.; Zhou, M.; et al. The terminal phycobilisome emitter, LCM: A light-harvesting pigment with a phytochrome chromophore. Proc. Natl. Acad. Sci. USA 2015, 112, 15880–15885. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.J.; Rockwell, N.C.; Jang, A.Y.; Ernst, L.A.; Waggoner, A.S.; Duan, Y.; Lei, H.; Lagarias, J.C. Multiple roles of a conserved GAF domain tyrosine residue in cyanobacterial and plant phytochromes. Biochemistry 2005, 44, 15203–15215. [Google Scholar] [CrossRef] [PubMed]
- Rockwell, N.C.; Shang, L.; Martin, S.S.; Lagarias, J.C. Distinct classes of red/far-red photochemistry within the phytochrome superfamily. Proc. Natl. Acad. Sci. USA 2009, 106, 6123–6127. [Google Scholar] [CrossRef] [PubMed]
- Bjorling, S.C.; Zhang, C.F.; Farrens, D.L.; Song, P.S.; Kliger, D.S. Time-resolved circular dichroism of native oat phytochrome photointermediates. J. Am. Chem. Soc. 1992, 114, 4581–4588. [Google Scholar] [CrossRef]
- Velazquez Escobar, F.; Hildebrandt, T.; Utesch, T.; Schmitt, F.J.; Seuffert, I.; Michael, N.; Schulz, C.; Mroginski, M.A.; Friedrich, T.; Hildebrandt, P. Structural parameters controlling the fluorescence properties of phytochromes. Biochemistry 2014, 53, 20–29. [Google Scholar] [CrossRef]
- Borucki, B.; von Stetten, D.; Seibeck, S.; Lamparter, T.; Michael, N.; Mroginski, M.A.; Otto, H.; Murgida, D.H.; Heyn, M.P.; Hildebrandt, P. Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. J. Biol. Chem. 2005, 280, 34358–34364. [Google Scholar] [CrossRef]
- Toh, K.C.; Stojkovic, E.A.; van Stokkum, I.H.; Moffat, K.; Kennis, J.T. Fluorescence quantum yield and photochemistry of bacteriophytochrome constructs. Phys. Chem Chem Phys. 2011, 13, 11985–11997. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Verkhusha, V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 2013, 10, 751–754. [Google Scholar] [CrossRef]
- Glazer, A.N.; Fang, S. Chromophore content of blue-green algal phycobiliproteins. J. Biol. Chem. 1973, 248, 659–662. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Turoverov, K.K.; Kuznetsova, I.M. Intrinsic fluorescence of actin. J. Fluoresc. 2003, 13, 41–57. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Biktashev, A.G.; Dorofeiuk, A.V.; Kuznetsova, I.M. A complex of apparatus and programs for the measurement of spectral, polarization and kinetic characteristics of fluorescence in solution. Tsitologiia 1998, 40, 806–817. [Google Scholar] [PubMed]
- O’Connor, D.V.; Phillips, D. Time-correlated Single Photon Counting; New York Academic Press: New York, NY, USA, 1984; pp. 37–54. [Google Scholar]
- Marquardt, D.W. An algorithm for least-squares estimation of non linear parameters. J. Soc. Ind. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Thümmler, F.; Brandlmeier, T.; Rüdiger, W. Preparation and Properties of Chromopeptides from the Pfr Form of Phytochrome. Z. Für Nat. C 1981, 36, 440–449. [Google Scholar] [CrossRef]
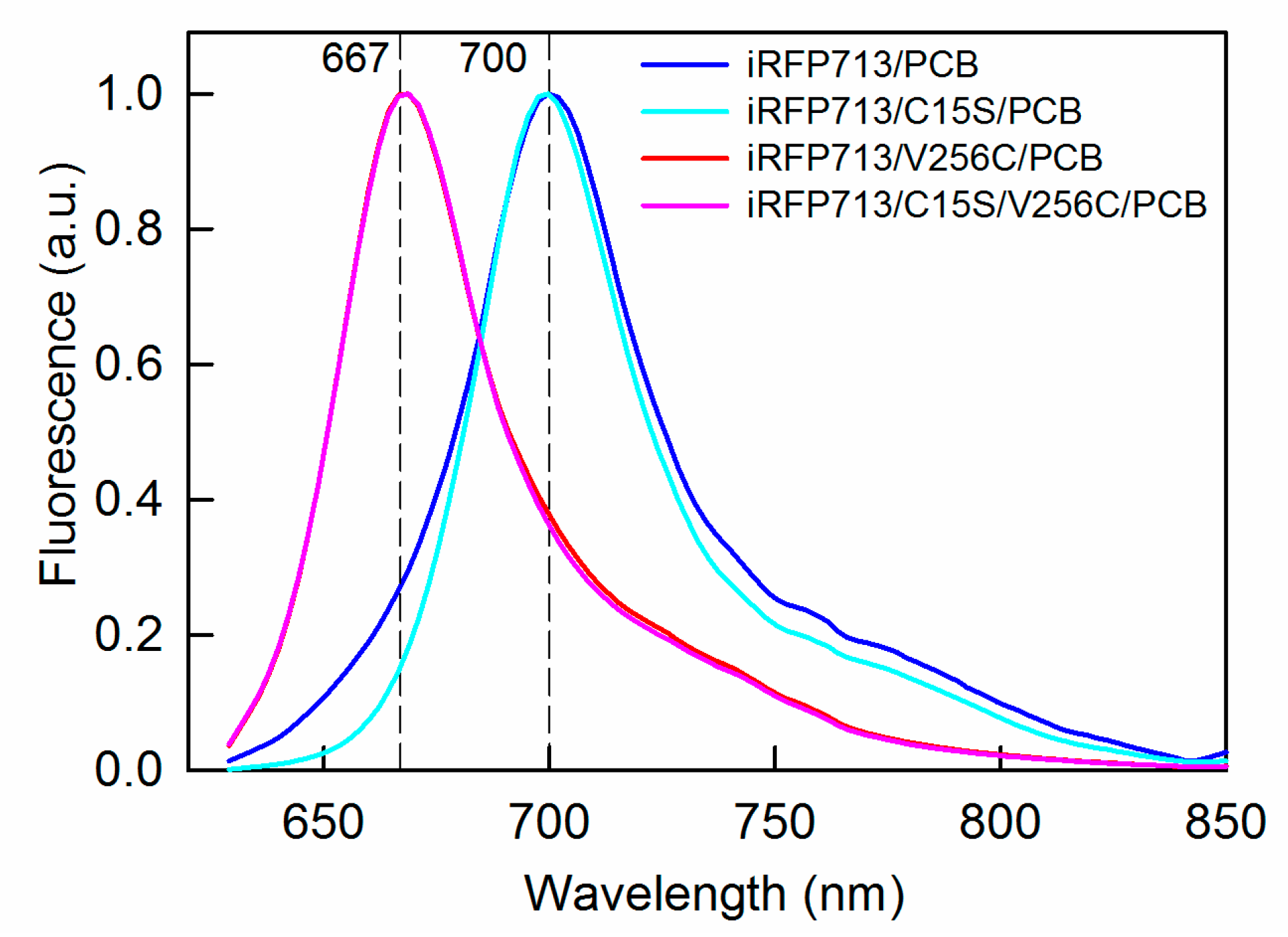
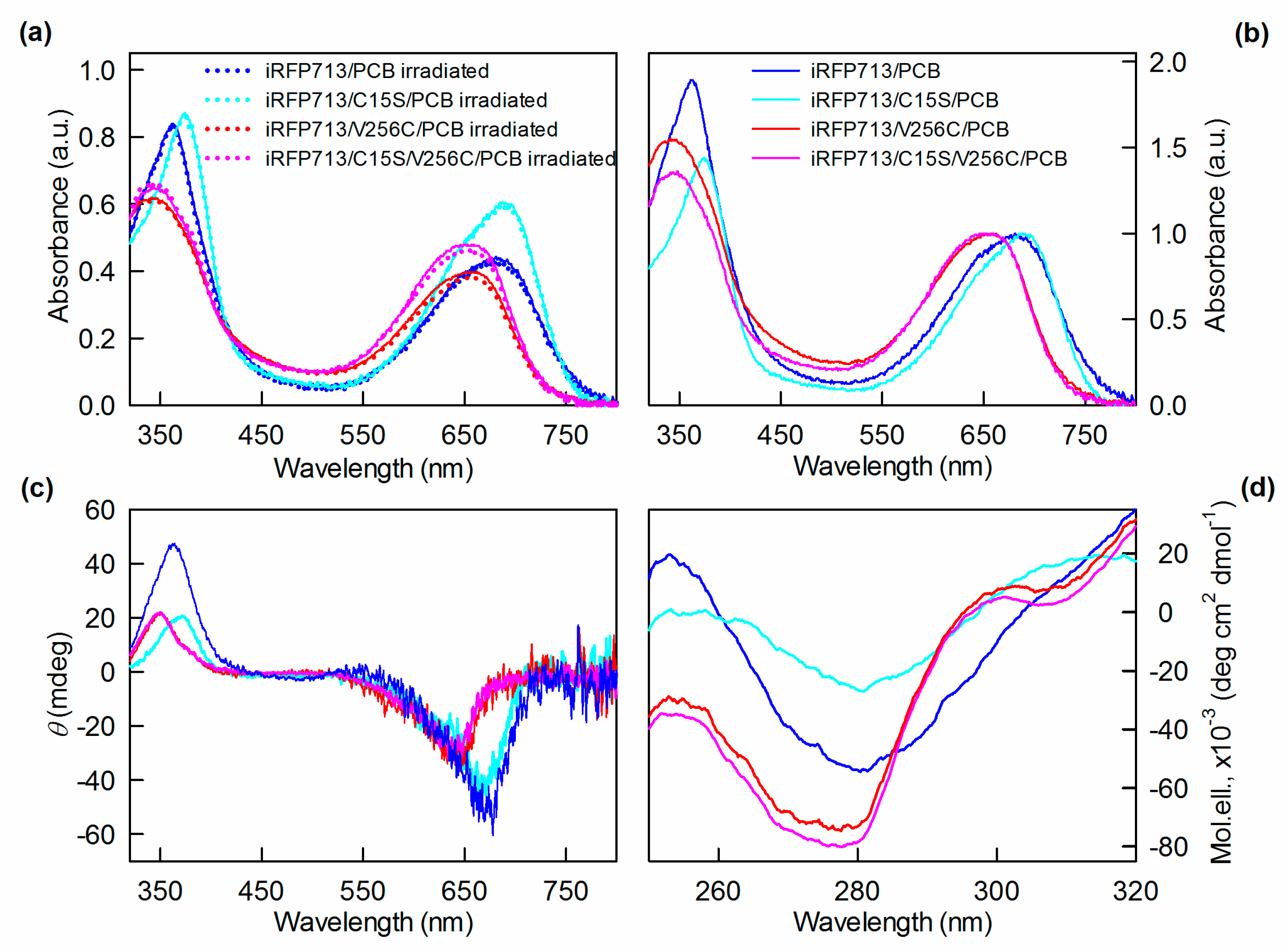
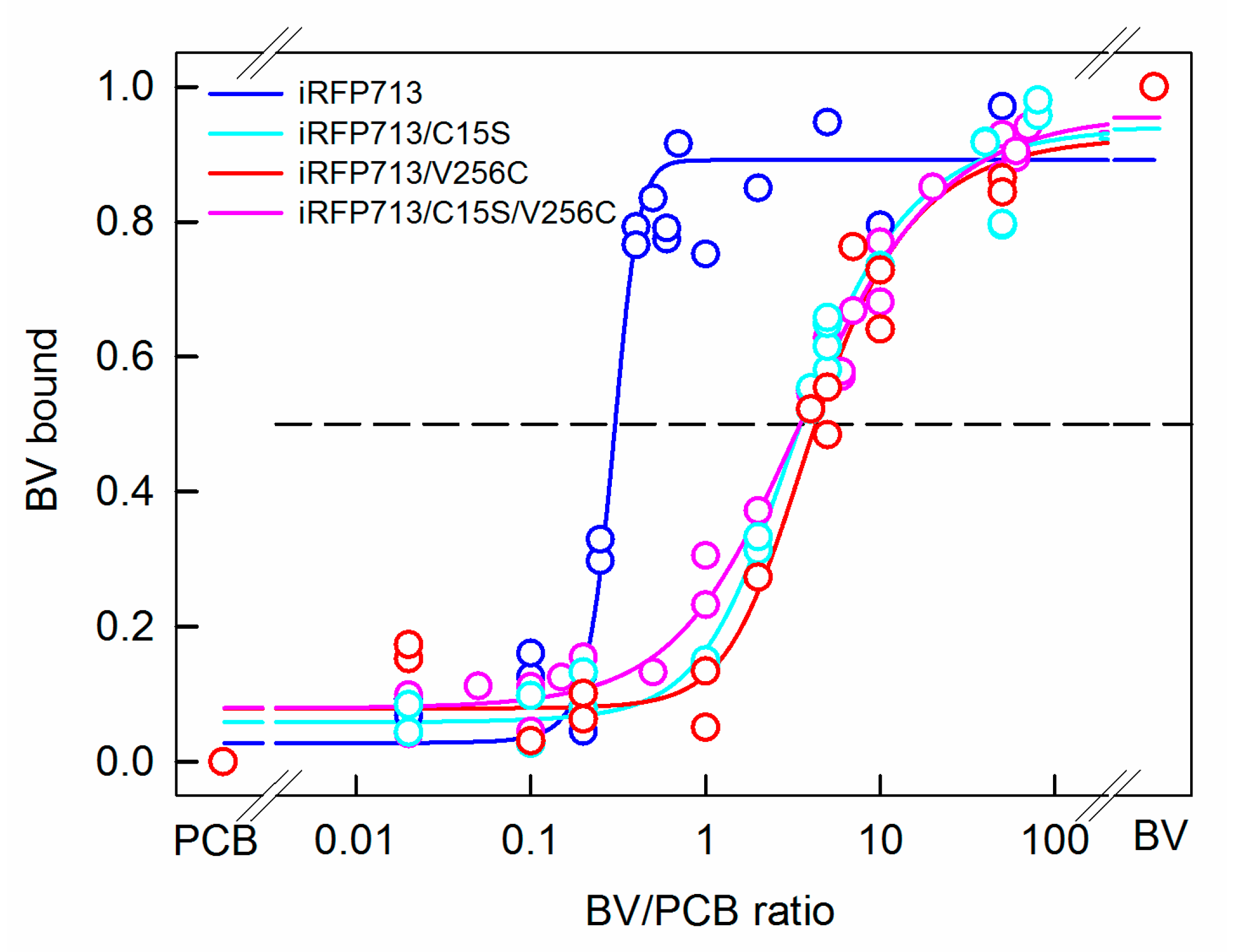
| Protein | Parameter A (λex = 295 nm) | Fl. Anisotropy r (λex = 295 nm, λem = 365 nm) | SAR 1 |
|---|---|---|---|
| iRFP713/BV | 1.56 ± 0.02 | 0.125 ± 0.005 | 1.9 |
| iRFP713/PCB | 1.73 ± 0.03 | 0.125 ± 0.005 | 1.8 |
| iRFP713/C15S/PCB | 1.70 ± 0.02 | 0.11 ± 0.01 | 2.0 |
| iRFP713/V256C/PCB | 1.65 ± 0.03 | 0.125 ± 0.005 | 1.9 |
| iRFP713/C15S/V256C/PCB | 1.64 ± 0.02 | 0.12 ± 0.01 | 2.0 |
| Protein | Helix 1 | β-Sheet | β-Turn | Random |
|---|---|---|---|---|
| iRFP713/BV | 0.19 | 0.31 | 0.20 | 0.29 |
| iRFP713/PCB | 0.19 | 0.31 | 0.21 | 0.29 |
| iRFP713/C15S/PCB | 0.18 | 0.30 | 0.21 | 0.31 |
| iRFP713/V256C/PCB | 0.18 | 0.31 | 0.21 | 0.31 |
| iRFP713/C15S/V256C/PCB | 0.18 | 0.30 | 0.22 | 0.30 |
| Protein | Absorbance Maximum (nm) | Extinction Coefficient at the Main Peak (M−1·cm−1) | Emission Maximum (nm) | Quantum Yield(%) | Chromophore Fluorescence Lifetime (ns) |
|---|---|---|---|---|---|
| iRFP713/BV | 692 ± 1 | 98,000 | 713 ± 1 | 6.3 1 | 0.67 ± 0.01 |
| iRFP713/C15S/BV | 685 ± 1 | 68,000 | 710 ± 1 | 5.5 ± 0.3 | 0.87 ± 0.01 |
| iRFP713/V256C/BV | 662 ± 1 | 94,000 | 680 ± 1 | 14.5 ± 0.5 | 1.53 ± 0.02 |
| iRFP713/C15S/V256C/BV | 665 ± 2 | 66,000 | 676 ± 2 | 7.2 ± 0.3 | 1.31 ± 0.02 |
| iRFP713/PCB | 675 ± 1 | 72,800 | 700 ± 2 | 8.5 ± 0.5 | 0.82 ± 0.03 |
| iRFP713/C15S/PCB | 674 ± 1 | 74,000 | 698 ± 1 | 17 ± 1.5 | 1.24 ± 0.02 |
| iRFP713/V256C/PCB | 646 ± 1 | 77,200 | 667 ± 1 | 45 ± 2.0 | 2.05 ± 0.01 |
| iRFP713/C15S/V256C/PCB | 647 ± 1 | 77,400 | 666 ± 1 | 50 ± 2.5 | 2.12 ± 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stepanenko, O.V.; Stepanenko, O.V.; Shpironok, O.G.; Fonin, A.V.; Kuznetsova, I.M.; Turoverov, K.K. Near-Infrared Markers based on Bacterial Phytochromes with Phycocyanobilin as a Chromophore. Int. J. Mol. Sci. 2019, 20, 6067. https://doi.org/10.3390/ijms20236067
Stepanenko OV, Stepanenko OV, Shpironok OG, Fonin AV, Kuznetsova IM, Turoverov KK. Near-Infrared Markers based on Bacterial Phytochromes with Phycocyanobilin as a Chromophore. International Journal of Molecular Sciences. 2019; 20(23):6067. https://doi.org/10.3390/ijms20236067
Chicago/Turabian StyleStepanenko, Olesya V., Olga V. Stepanenko, Olesya G. Shpironok, Alexander V. Fonin, Irina M. Kuznetsova, and Konstantin K. Turoverov. 2019. "Near-Infrared Markers based on Bacterial Phytochromes with Phycocyanobilin as a Chromophore" International Journal of Molecular Sciences 20, no. 23: 6067. https://doi.org/10.3390/ijms20236067
APA StyleStepanenko, O. V., Stepanenko, O. V., Shpironok, O. G., Fonin, A. V., Kuznetsova, I. M., & Turoverov, K. K. (2019). Near-Infrared Markers based on Bacterial Phytochromes with Phycocyanobilin as a Chromophore. International Journal of Molecular Sciences, 20(23), 6067. https://doi.org/10.3390/ijms20236067








