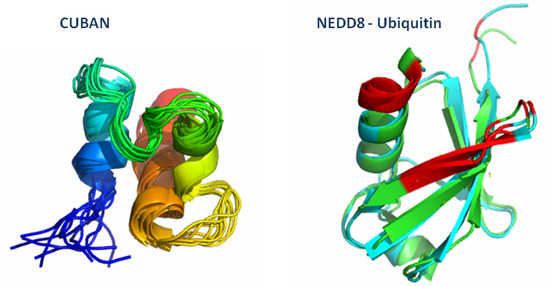
CUBAN, a Case Study of Selective Binding: Structural Details of the Discrimination between Ubiquitin and NEDD8
Abstract
1. Introduction
2. Results
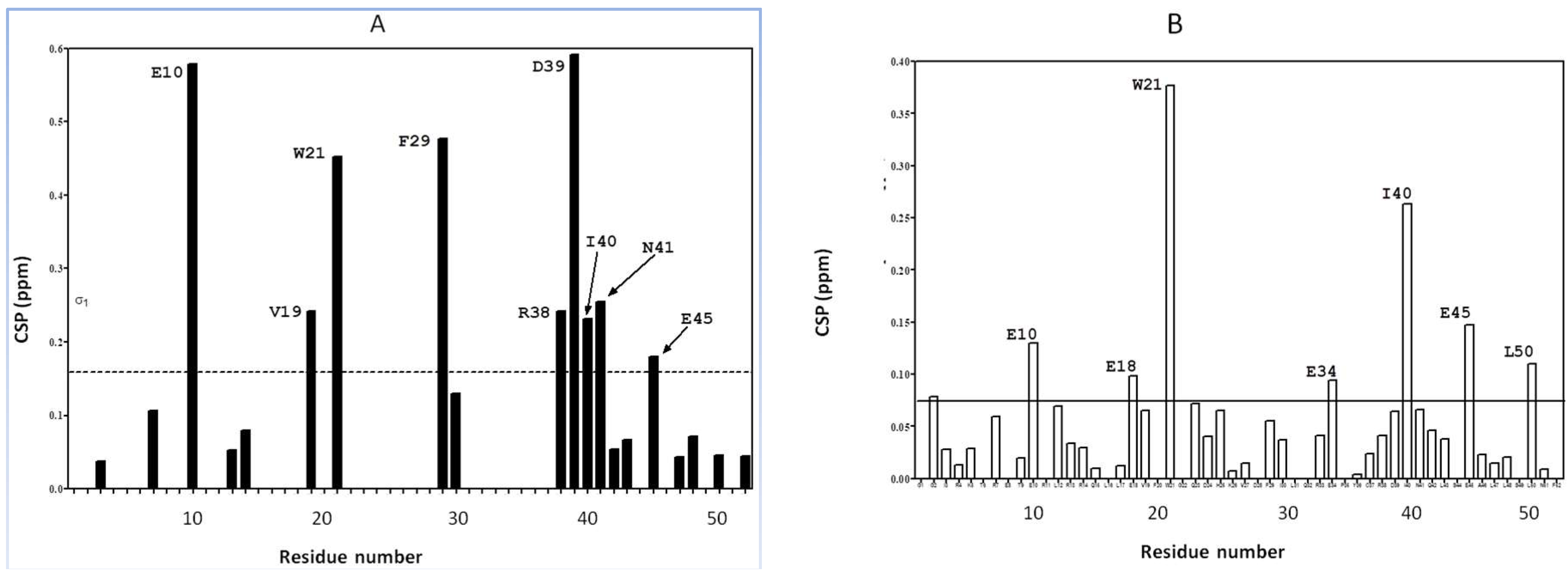
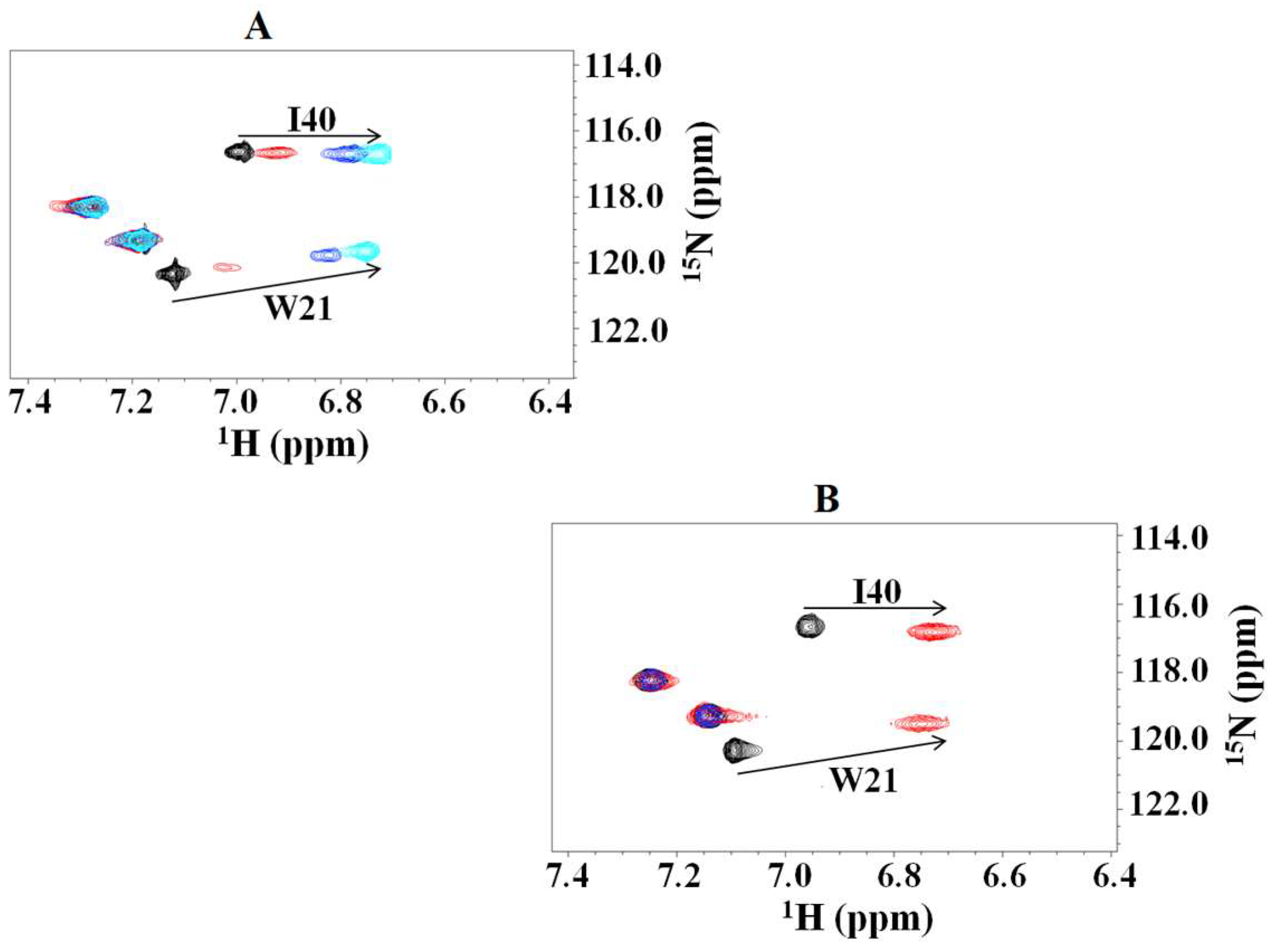
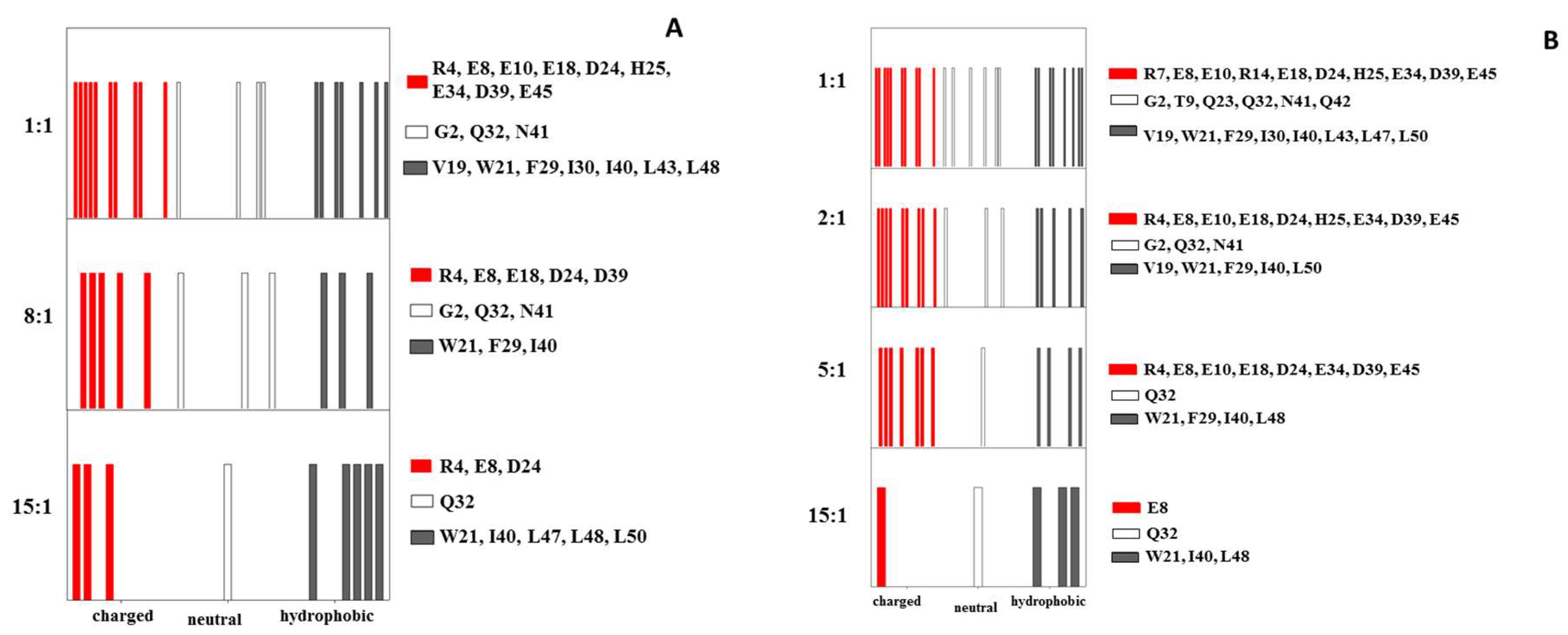
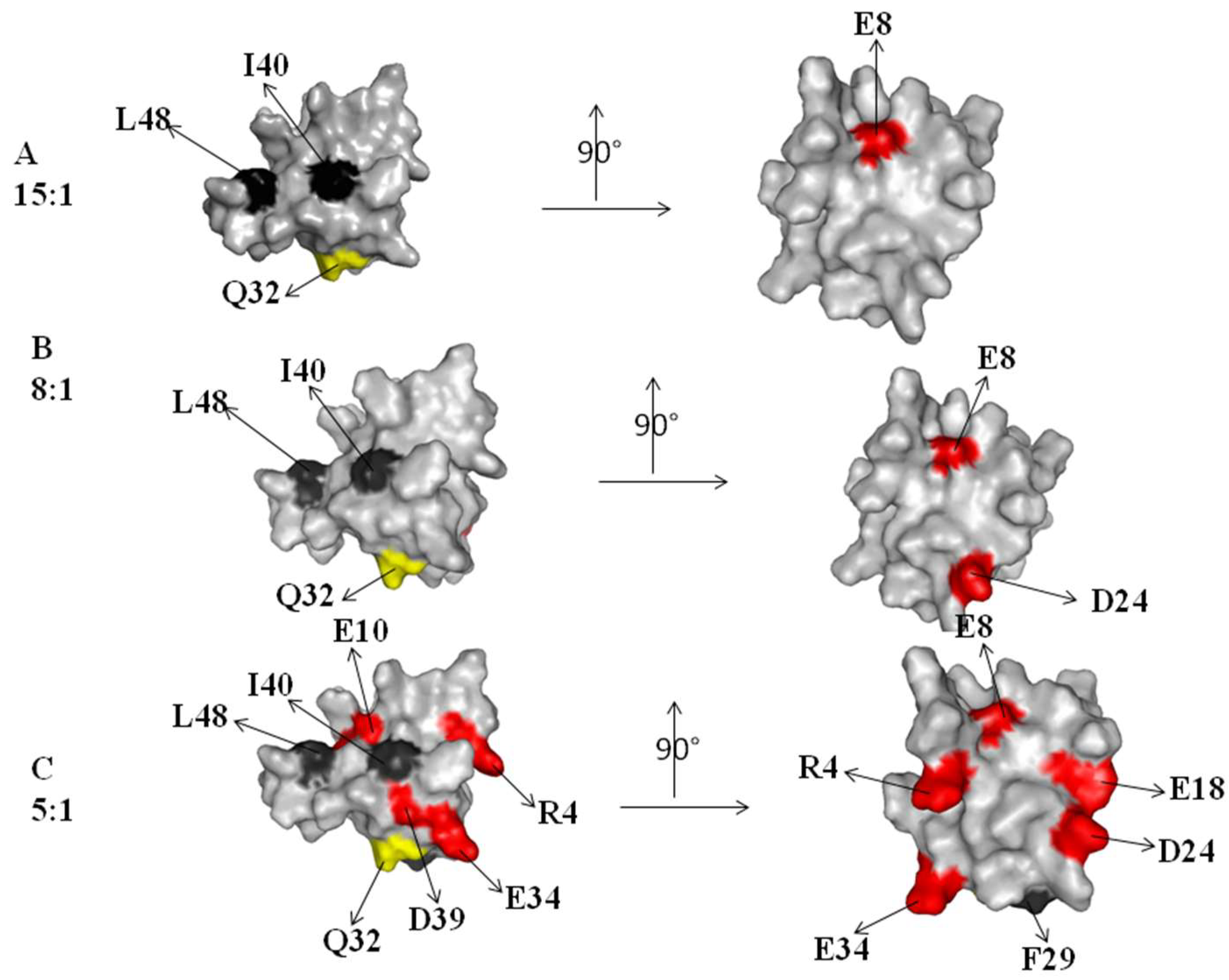
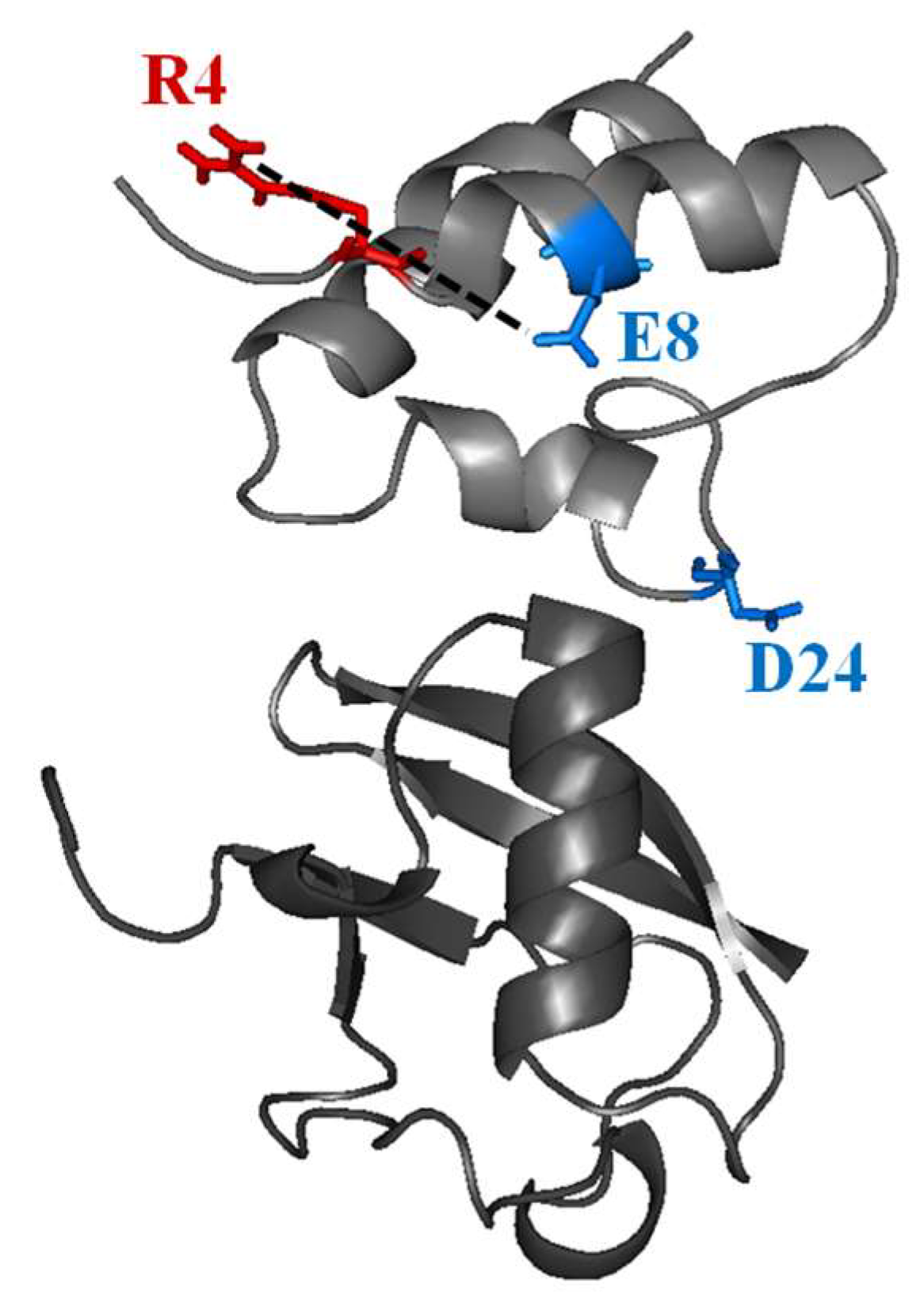
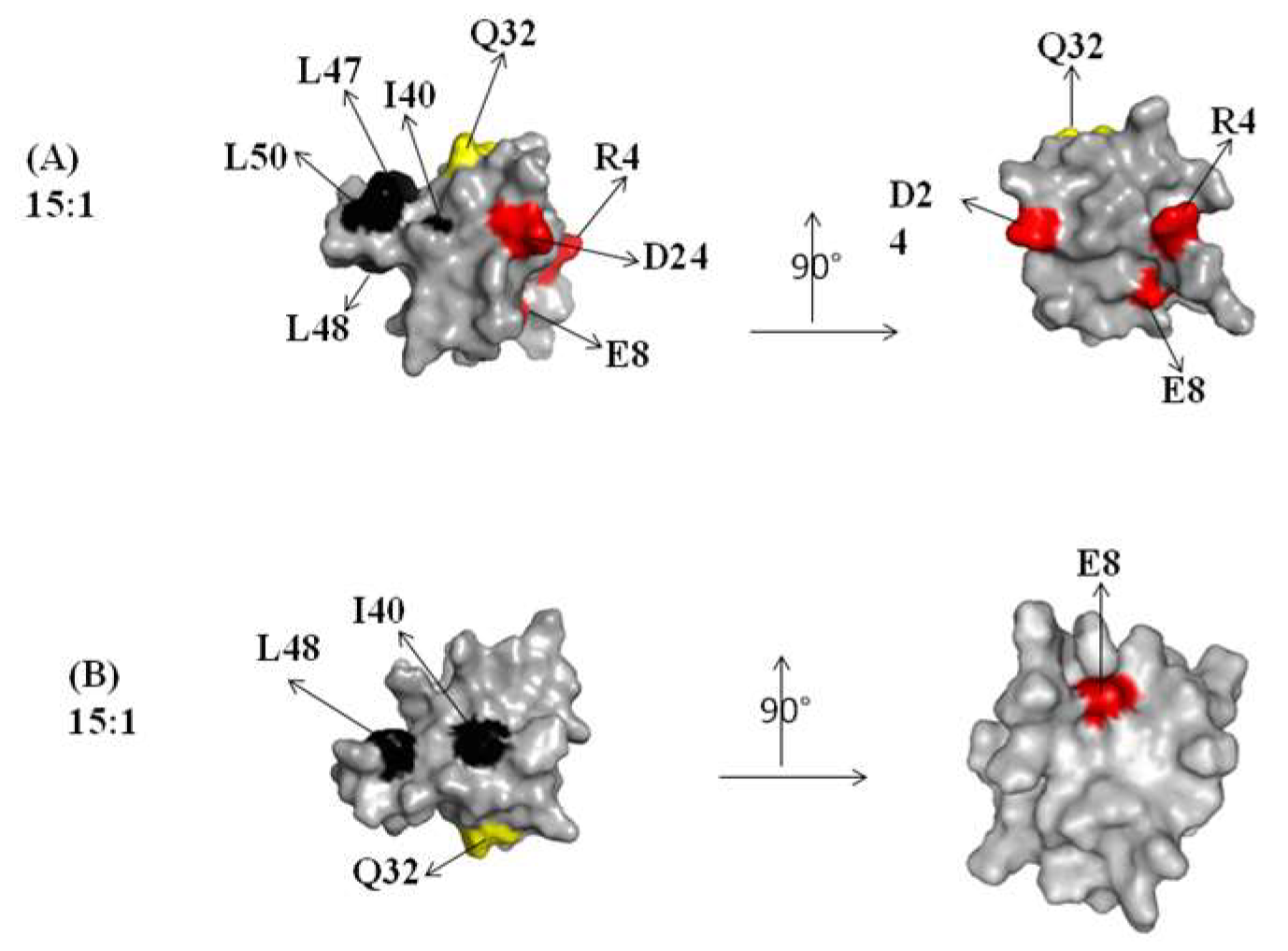
2.1. Interaction of the 15N Labeled CUBAN Domain with NEDD8 and Ubiquitin
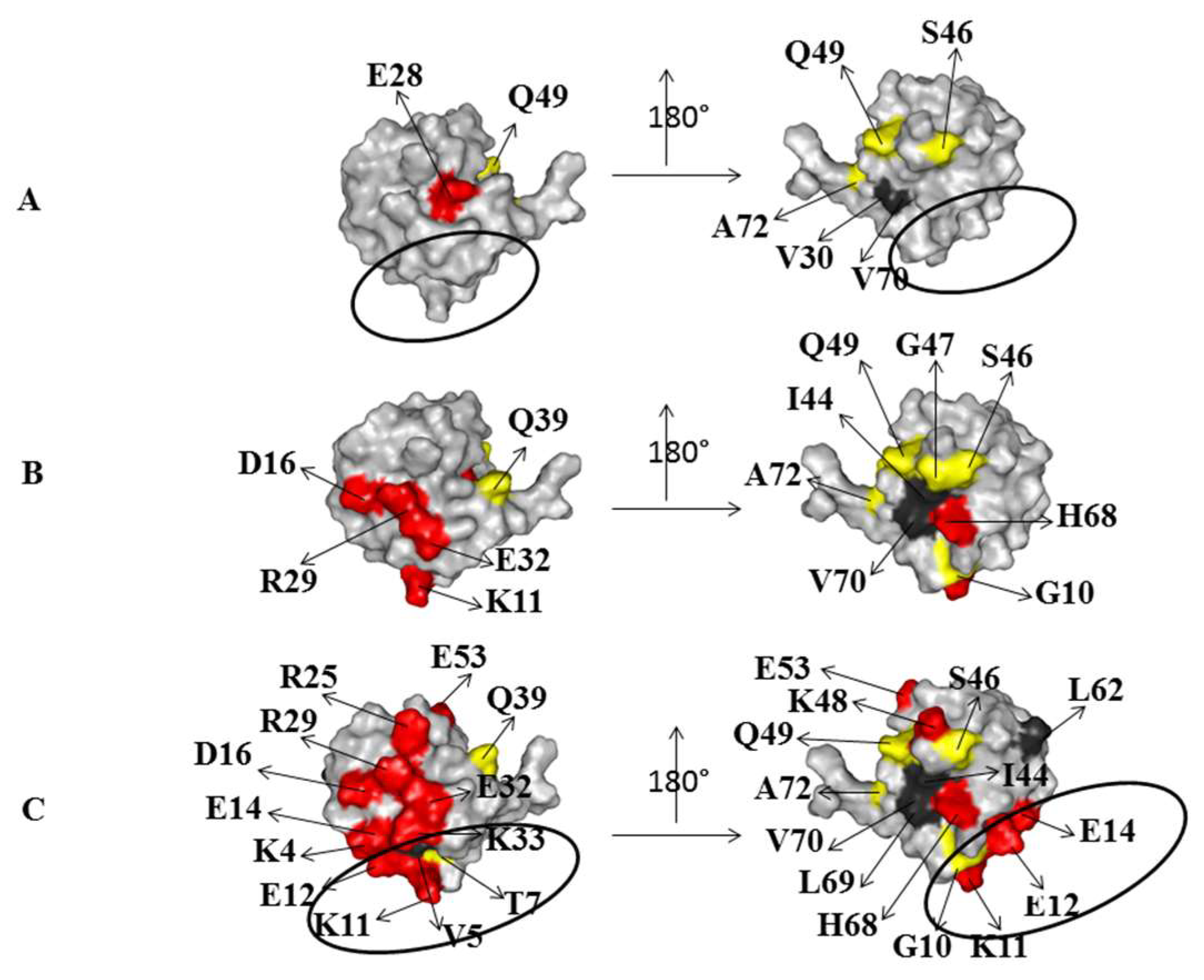
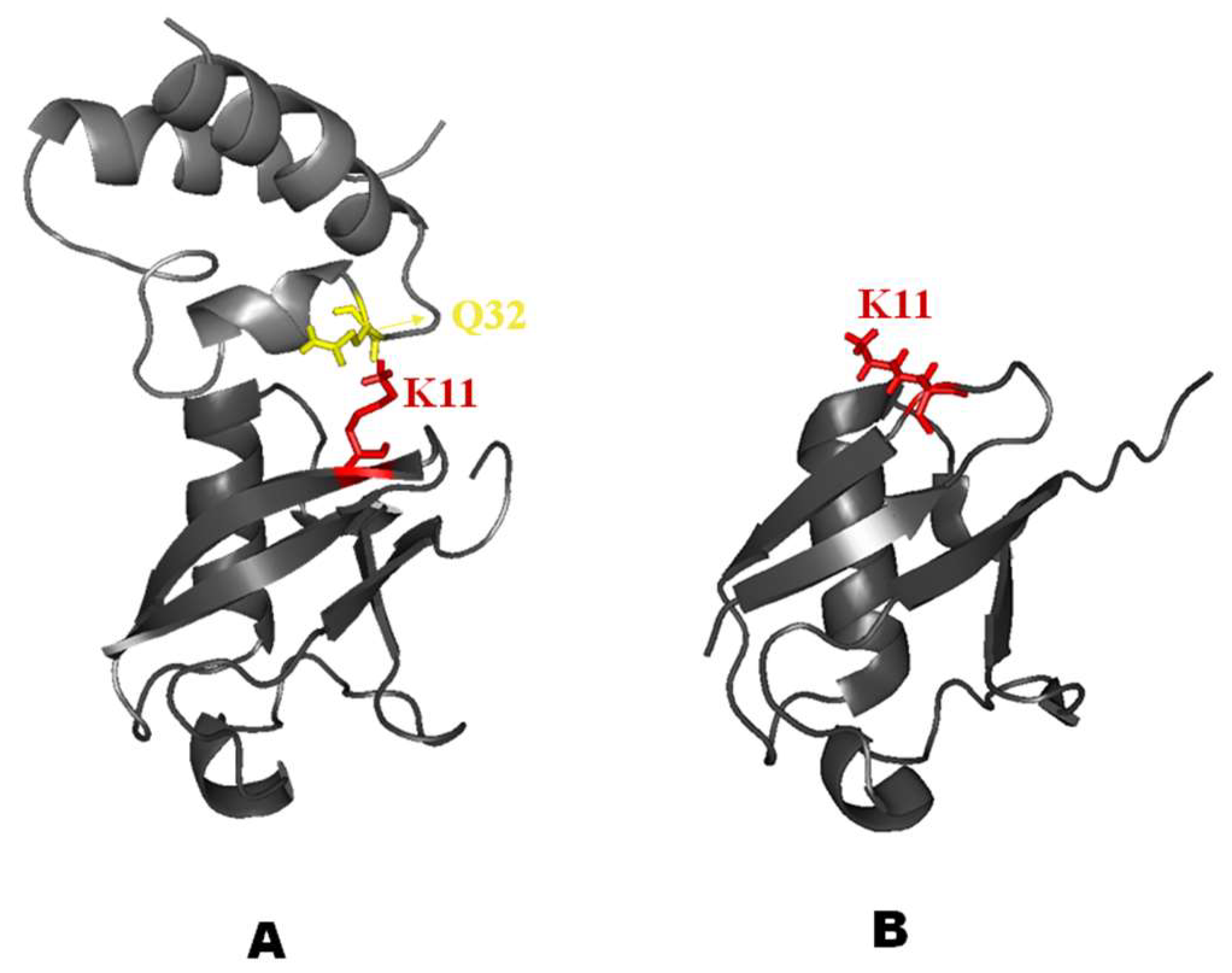
2.2. Interaction of 15N Labeled NEDD8 with the CUBAN Domain
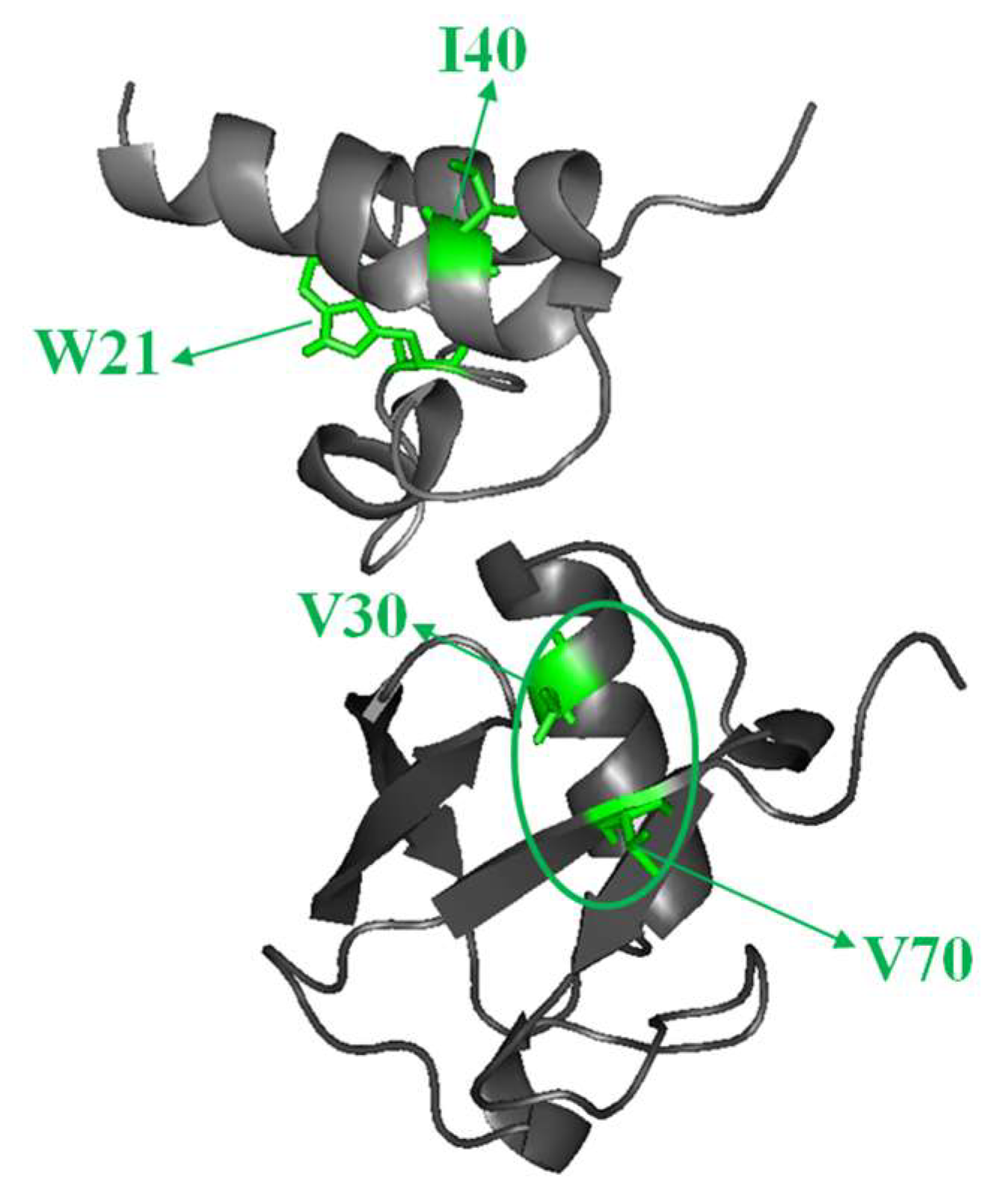
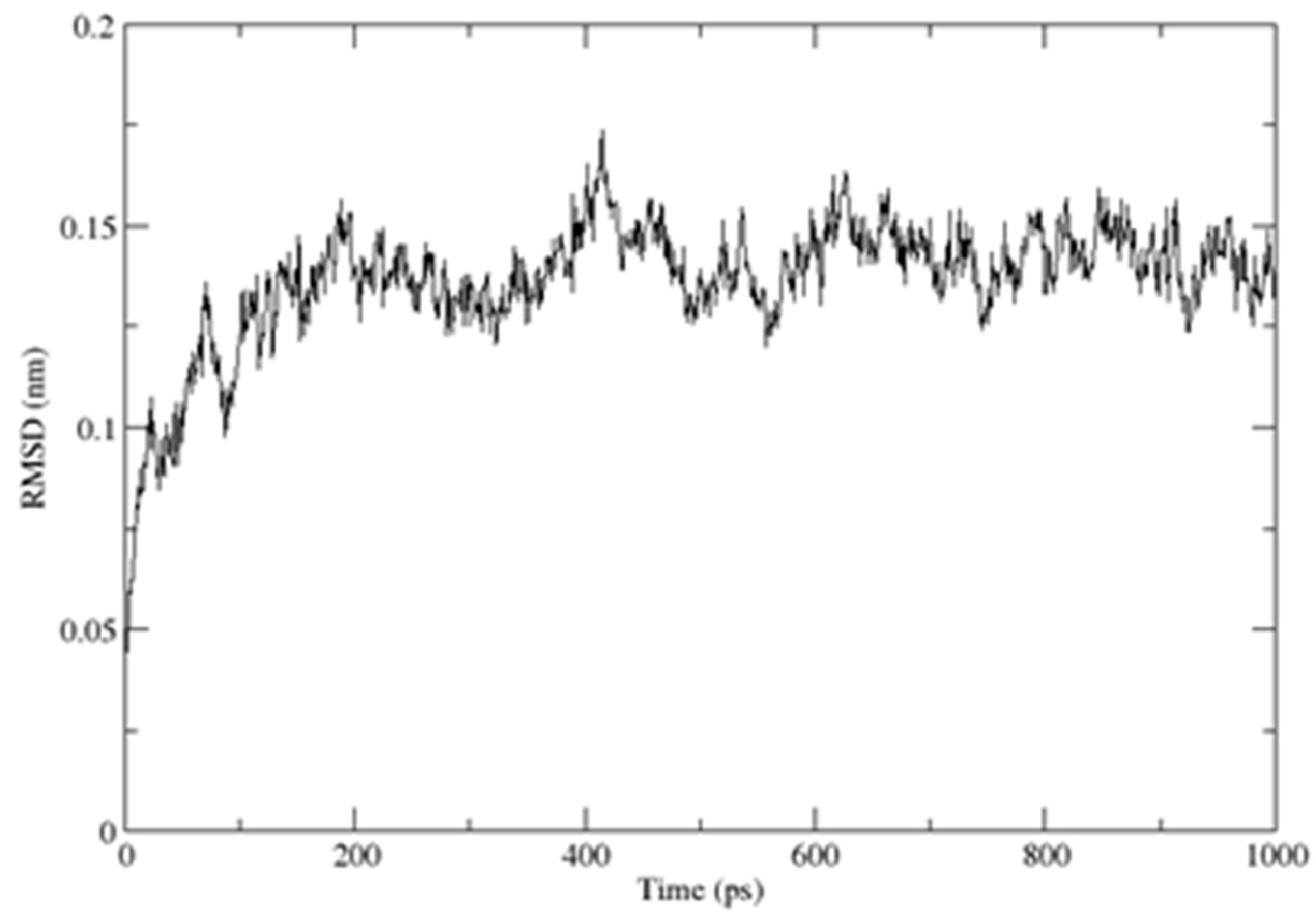
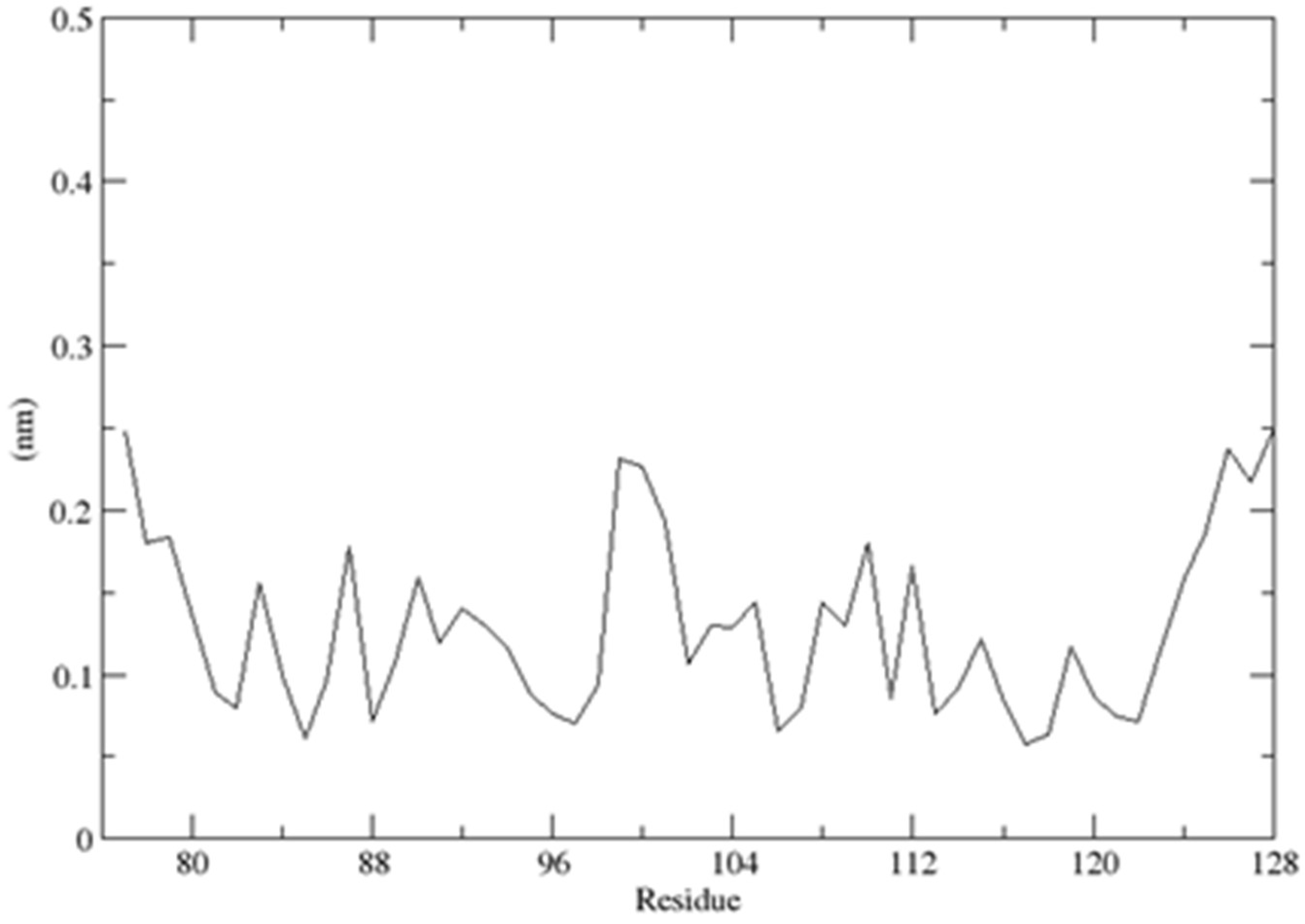
2.3. Structural Dynamics of the CUBAN Domain in Complex with NEDD8
3. Discussion
Conformational Selection or Induced Fit?
4. Materials and Methods
4.1. Expression Plasmids and Protein Purification
4.2. NMR Spectroscopy
4.3. Molecular Dynamics Simulations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Mayer, R.J.; Landon, M.; Layfield, R. Ubiquitin superfolds: Intrinsic and attachable regulators of cellular activities? Fold. Des. 1998, 3, R97–R99. [Google Scholar] [CrossRef]
- Huang, D.T.; Miller, D.W.; Mathew, R.; Cassell, R.; Holton, J.M.; Roussel, M.F.; Schulman, B.A. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 2004, 11, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.T.; Paydar, A.; Zhuang, M.; Waddell, M.B.; Holton, J.M.; Schulman, B.A. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol. Cell 2005, 17, 341–350. [Google Scholar] [CrossRef]
- Huang, D.T.; Hunt, H.W.; Zhuang, M.; Ohi, M.D.; Holton, J.M.; Schulman, B.A. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature 2007, 445, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Oved, S.; Mosesson, Y.; Zwang, Y.; Santonico, E.; Shtiegman, K.; Marmor, M.D.; Kochupurakkal, B.S.; Katz, M.; Lavi, S.; Cesareni, G.; et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 2006, 281, 21640–21651. [Google Scholar] [CrossRef]
- Den Besten, W.; Verma, R.; Kleiger, G.; Oania, R.S.; Deshaies, R.J. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat. Struct. Mol. Biol. 2012, 19, 511–516. [Google Scholar] [CrossRef]
- Bandau, S.; Knebel, A.; Gage, Z.O.; Wood, N.T.; Alexandru, G. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation. BMC Biol. 2012, 10, 36. [Google Scholar] [CrossRef]
- Kelsall, I.R.; Duda, D.M.; Olszewski, J.L.; Hofmann, K.; Knebel, A.; Langevin, F.; Wood, N.; Wightman, M.; Schulman, B.A.; Alpi, A.F. TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 2013, 32, 2848–2860. [Google Scholar] [CrossRef]
- Tanaka, T.; Kawashima, H.; Yeh, E.T.H.; Kamitani, T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J. Biol. Chem. 2003, 278, 32905–32913. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, G.; Kalveram, B.; Weber, E.; Bochtler, P.; Lukasiak, S.; Hipp, M.S.; Groettrup, M. The UBA domains of NUB1L are required for binding but not for accelerated degradation of the ubiquitin-like modifier FAT10. J. Biol. Chem. 2006, 281, 20045–20054. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Chen, Y.; Zhang, F.; Yang, C.-Y.; Wang, S.; Yu, X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell 2013, 49, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Castagnoli, L.; Mandaliti, W.; Nepravishta, R.; Valentini, E.; Mattioni, A.; Procopio, R.; Iannuccelli, M.; Polo, S.; Paci, M.; Cesareni, G. Santonico Selectivity of the CUBAN domain in the recognition of ubiquitin and NEDD8. FEBS J. 2019, 98, 1–37. [Google Scholar]
- Sundd, M.; Iverson, N.; Ibarra-Molero, B.; Sanchez-Ruiz, J.M.; Robertson, A.D. Electrostatic interactions in ubiquitin: Stabilization of carboxylates by lysine amino groups. Biochemistry 2002, 41, 7586–7596. [Google Scholar] [CrossRef] [PubMed]
- Wlodarski, T.; Zagrovic, B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. USA 2009, 106, 19346–19351. [Google Scholar] [CrossRef] [PubMed]
- Lange, O.F.; Lakomek, N.-A.; Farès, C.; Schröder, G.F.; Walter, K.F.A.; Becker, S.; Meiler, J.; Grubmüller, H.; Griesinger, C.; de Groot, B.L. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science 2008, 320, 1471–1475. [Google Scholar] [CrossRef]
- Changeux, J.-P.; Edelstein, S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 2011, 3, 19. [Google Scholar] [CrossRef]
- Kurauskas, V.; Izmailov, S.A.; Rogacheva, O.N.; Hessel, A.; Ayala, I.; Woodhouse, J.; Shilova, A.; Xue, Y.; Yuwen, T.; Coquelle, N.; et al. Slow conformational exchange and overall rocking motion in ubiquitin protein crystals. Nat. Commun. 2017, 8, 145. [Google Scholar] [CrossRef]
- Hammes, G.G.; Chang, Y.-C.; Oas, T.G. Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA 2009, 106, 13737–13741. [Google Scholar] [CrossRef]
- Peters, J.H.; de Groot, B.L. Ubiquitin dynamics in complexes reveal molecular recognition mechanisms beyond induced fit and conformational selection. PLoS Comput. Biol. 2012, 8, e1002704. [Google Scholar] [CrossRef] [PubMed]
- Weber, D.J.; Gittis, A.G.; Mullen, G.P.; Abeygunawardana, C.; Lattman, E.E.; Mildvan, A.S. NMR docking of a substrate into the X-ray structure of staphylococcal nuclease. Proteins 1992, 13, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Bodenhausen, G.; Riben, J.D. Natural abundance Nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 2001, 69, 185–189. [Google Scholar] [CrossRef]
- Nguyen, B.D.; Meng, X.; Donovan, K.J.; Shaka, A.J. SOGGY: Solvent-optimized double gradient spectroscopy for water suppression. A comparison with some existing techniques. J. Magn. Reson. 2007, 184, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Zuiderweg, E.R.P. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry 2002, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santonico, E.; Nepravishta, R.; Mandaliti, W.; Castagnoli, L.; Cesareni, G.; Paci, M. CUBAN, a Case Study of Selective Binding: Structural Details of the Discrimination between Ubiquitin and NEDD8. Int. J. Mol. Sci. 2019, 20, 1185. https://doi.org/10.3390/ijms20051185
Santonico E, Nepravishta R, Mandaliti W, Castagnoli L, Cesareni G, Paci M. CUBAN, a Case Study of Selective Binding: Structural Details of the Discrimination between Ubiquitin and NEDD8. International Journal of Molecular Sciences. 2019; 20(5):1185. https://doi.org/10.3390/ijms20051185
Chicago/Turabian StyleSantonico, Elena, Ridvan Nepravishta, Walter Mandaliti, Luisa Castagnoli, Gianni Cesareni, and Maurizio Paci. 2019. "CUBAN, a Case Study of Selective Binding: Structural Details of the Discrimination between Ubiquitin and NEDD8" International Journal of Molecular Sciences 20, no. 5: 1185. https://doi.org/10.3390/ijms20051185
APA StyleSantonico, E., Nepravishta, R., Mandaliti, W., Castagnoli, L., Cesareni, G., & Paci, M. (2019). CUBAN, a Case Study of Selective Binding: Structural Details of the Discrimination between Ubiquitin and NEDD8. International Journal of Molecular Sciences, 20(5), 1185. https://doi.org/10.3390/ijms20051185