- Article
Reliable Gene Expression Normalization in Cucumber Leaves: Identifying Stable Reference Genes Under Drought Stress
- Wojciech Szczechura,
- Urszula Kłosińska and
- Marzena Nowakowska
- + 2 authors
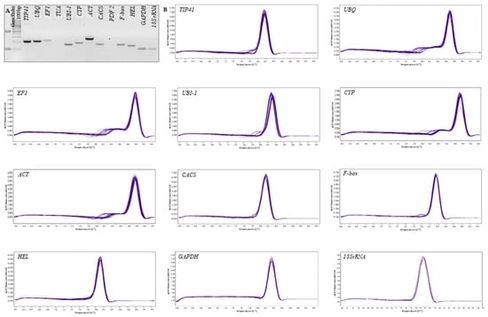
Reverse transcription quantitative PCR (RT-qPCR) is extensively used to quantify gene expression under drought conditions; however, its reliability depends on the validation of the reference genes under specific conditions. In cucumber, reference genes have rarely been validated under drought conditions. This study identified stable housekeeping genes for RT-qPCR normalization in the leaves of two inbred lines with contrasting drought responses. Plants underwent a 7-day drought period, with leaf samples collected at multiple points along with watered controls. The expression stability of 13 candidate genes was evaluated using four algorithms: geNorm, NormFinder, BestKeeper, and the comparative ΔCt method, with the results integrated using RefFinder. Ten genes producing specific and efficient amplicons were analyzed for stability. CACS and UBI-1 consistently ranked among the most stable genes, with TIP41-like as an additional reliable option, whereas GAPDH and HEL were unstable. GeNorm pairwise variation analysis showed that the two reference genes were sufficient for accurate normalization. Functional validation with three drought-responsive targets (LOX, HsfC1, and CYP72A219) and comparison with RNA sequencing (RNA-seq) fold changes confirmed that normalization using CACS and UBI-1 yielded the most biologically credible expression profiles. These reference genes will facilitate robust RT-qPCR analyses of drought response in cucumber leaves and provide a starting point for validating suitable normalizers in other cucumber organs and related cucurbits.
6 December 2025