Journal Description
Microorganisms
Microorganisms
is a scientific, peer-reviewed, open access journal of microbiology, published monthly online by MDPI. The Hellenic Society Mikrobiokosmos (MBK), the Spanish Society for Nitrogen Fixation (SEFIN) and the Society for Microbial Ecology and Disease (SOMED) are affiliated with Microorganisms, and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, PubAg, CAPlus / SciFinder, AGRIS, and other databases.
- Journal Rank: JCR - Q2 (Microbiology) / CiteScore - Q1 (Microbiology (medical))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 15.2 days after submission; acceptance to publication is undertaken in 2.9 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Testimonials: See what our editors and authors say about Microorganisms.
- Companion journal for Microorganisms include: Applied Microbiology and Bacteria.
Impact Factor:
4.2 (2024);
5-Year Impact Factor:
4.6 (2024)
Latest Articles
Unraveling the Potential Antiviral Activity of Isoxazoline-Carbocyclic Monophosphate Nucleotides Against Echovirus 11
Microorganisms 2025, 13(12), 2662; https://doi.org/10.3390/microorganisms13122662 (registering DOI) - 23 Nov 2025
Abstract
From July 2022, a novel Echovirus 11 (E11) variant has been associated with severe neonatal infections and liver failure. Currently, there are no vaccines or antiviral options for the targeted treatment of non-polio enterovirus (EV) infections; therefore, anti-EV drugs are urgently needed. In
[...] Read more.
From July 2022, a novel Echovirus 11 (E11) variant has been associated with severe neonatal infections and liver failure. Currently, there are no vaccines or antiviral options for the targeted treatment of non-polio enterovirus (EV) infections; therefore, anti-EV drugs are urgently needed. In this study, the putative anti-E11 activity of two isoxzoline-carbocyclic monophosphate nucleotides (4a and 4b) was evaluated in vitro by cytopathic effect (CPE) reduction in VERO 76 cells and qRT-PCR. Treatment with nucleotide 4a at 25 and 50 μM successfully diminished the CPE caused by E11 by 90% and 75%, respectively, and induced a reduction in viral RNA in the supernatant by 72% and 89%. In contrast, the treatment with 25 and 50 μM of 4b caused a minor inhibition of CPE (58 and 38%), and no significant E11 RNA level changes were observed. A time course viral progeny production assay was performed to assess the inhibitory effect of nucleotide 4a on E11 infection progression. Compared to the control, the treated group showed a significant drop in viral RNA levels, with reductions of 43% at 10 h, 65% at 24 h, and 96% at 48 h post-infection. The results showed the extensive antiviral properties of the monophosphate nucleotide 4a in vitro. Moreover, a retrospective molecular docking study strongly supports that nucleotide 4a is an RdRp inhibitor capable of decreasing E11 genome replication and virus particle formation in VERO 76 cells.
Full article
(This article belongs to the Section Virology)
►
Show Figures
Open AccessArticle
Youngimonas ophiurae sp. nov., a Quorum-Quenching Marine Bacterium Isolated from a Brittle Star in the South China Sea, and Reclassification of Lutimaribacter litoralis as Youngimonas litoralis comb. nov
by
Zengzhi Liu, Meng Zhang, Qiliang Lai, Shanshan Xu and Ying Xu
Microorganisms 2025, 13(12), 2661; https://doi.org/10.3390/microorganisms13122661 (registering DOI) - 22 Nov 2025
Abstract
Two novel bacterial strains, designated S70T and S69A, were isolated from a marine brittle star collected in the South China Sea. These strains are Gram-stain-negative, non-motile, aerobic, and rod-shaped. A phylogenomic analysis indicated that strains S70T and S69A formed a distinct
[...] Read more.
Two novel bacterial strains, designated S70T and S69A, were isolated from a marine brittle star collected in the South China Sea. These strains are Gram-stain-negative, non-motile, aerobic, and rod-shaped. A phylogenomic analysis indicated that strains S70T and S69A formed a distinct branch with Youngimonas vesicularis CC-AMW-ET and Lutimaribacter litoralis JCM 17792T. The DNA G+C content of both strains was 61.5%. The digital DNA–DNA hybridization values with the closest relatives were 21.8, and 21.2%, respectively. Furthermore, the average nucleotide identity (ANIb) values between strain S70T and these two reference strains were 74.9% and 74.6%, respectively, both well below the 95–96% threshold for dividing prokaryotic species. The major fatty acids of strain S70T were summed feature 8 (C18:1 ω6c and/or C18:1 ω7c). Functional genomic analysis revealed that strain S70T possesses potential for hydrocarbon degradation and may play a significant role in sulfur metabolism. Additionally, strain S70T exhibited broad-spectrum AHL-degrading activity and, most notably, significantly inhibited soft rot caused by Pectobacterium carotovorum in potato tuber assays. Genomic comparisons further support the reclassification of Lutimaribacter litoralis into the genus Youngimonas.
Full article
(This article belongs to the Section Environmental Microbiology)
Open AccessArticle
Influence Pattern and Mechanism of Increased Nitrogen Deposition and AM Fungi on Soil Microbial Community in Desert Ecosystems
by
Hui Wang, Wan Duan, Qianqian Dong, Zhanquan Ji, Wenli Cao, Fangwei Zhang, Wenshuo Li and Yangyang Jia
Microorganisms 2025, 13(12), 2660; https://doi.org/10.3390/microorganisms13122660 (registering DOI) - 22 Nov 2025
Abstract
With continuous increases in nitrogen (N) deposition in the future, its impacts on terrestrial ecosystems are attracting growing concern. Arbuscular mycorrhiza (AM) fungi play a crucial role in shaping both soil microbial and plant communities. AM fungi play a crucial role in shaping
[...] Read more.
With continuous increases in nitrogen (N) deposition in the future, its impacts on terrestrial ecosystems are attracting growing concern. Arbuscular mycorrhiza (AM) fungi play a crucial role in shaping both soil microbial and plant communities. AM fungi play a crucial role in shaping the soil microbial and plant communities, yet their patterns of influence under increased N deposition scenarios remain unclear, particularly in desert ecosystems. Therefore, we conducted a field experiment simulating increased N deposition and AM fungal suppression to assess the effects of increased N deposition and AM fungi on soil microbial communities, employing phospholipid fatty acid (PLFA) biomarker technology in the Gurbantunggut Desert of Xinjiang. We found that increased N deposition promoted soil microbial biomass, including AM fungi, fungi, Actinomycetes (Act), Gram-positive bacteria (G+), Gram-negative bacteria (G−), and Dark Septate Endophyte (DSE). AM fungal suppression significantly increased the content of soil Act and G+. There were clearly and significantly interactive effects of increased N deposition and AM fungi on soil microbial contents. Both increased N deposition and AM fungi caused significant changes in soil microbial community structure. Random forest analysis revealed that soil nitrate N (NO3−-N), Soil Organic Carbon (SOC), and pH were main factors influencing soil microorganisms; soil AM fungi, G+, and Act significantly affected plant Shannon diversity; soil G−, Act, and fungi posed significant effects on plant community biomass. Finally, the structure equation model results indicated that soil fungi, especially AM fungi, were the main soil microorganisms altering the plant community diversity and biomass under increased N deposition. This study reveals the crucial role of AM fungi in regulating soil microbial responses to increased N deposition, providing experimental evidence for understanding how N deposition affects plant communities through soil microorganisms.
Full article
(This article belongs to the Special Issue Soil Health and Plant Microbiome–Bioeffectors Relationship in Sustainable Agriculture, 2nd Edition)
Open AccessArticle
Gut Microbiota-Targeted Photobiomodulation Ameliorates Alzheimer’s Pathology via the Gut–Brain Axis: Comparable Efficacy to Transcranial Irradiation
by
Shisheng Cao, Xinyu Shi, Yongqiang Chen, Tiaotiao Liu, Jiashen Hu, Xiaoxi Dong, Hongli Chen, Jianwu Dai and Huijuan Yin
Microorganisms 2025, 13(12), 2659; https://doi.org/10.3390/microorganisms13122659 (registering DOI) - 22 Nov 2025
Abstract
Alzheimer’s disease (AD) is a major neurodegenerative disorder with limited effective and affordable therapies. Photobiomodulation (PBM) offers a safe, non-invasive treatment strategy, yet conventional transcranial PBM (tc-PBM) is restricted by low skull penetration. To overcome this limitation, gut microbiota-targeted PBM (gm-PBM) has been
[...] Read more.
Alzheimer’s disease (AD) is a major neurodegenerative disorder with limited effective and affordable therapies. Photobiomodulation (PBM) offers a safe, non-invasive treatment strategy, yet conventional transcranial PBM (tc-PBM) is restricted by low skull penetration. To overcome this limitation, gut microbiota-targeted PBM (gm-PBM) has been proposed to modulate the gut–brain axis, though its efficacy and mechanisms remain unclear. Here, six-month-old APPswe/PS1dE9 mice received gm-PBM or tc-PBM (810 nm, 25 mW/cm2, 20 min/day for 4 weeks). Behavioral testing revealed that both treatments improved spatial learning and memory, while histological analyses showed reduced amyloid-β deposition and microglial shift toward an anti-inflammatory phenotype. Notably, gm-PBM specifically enriched short-chain fatty acid-producing bacteria, elevated propionate, butyrate, and secondary bile acids, and restored intestinal barrier integrity, whereas tc-PBM induced minimal microbiota changes. These findings suggest that gm-PBM confers neuroprotective effects comparable to or exceeding tc-PBM through modulation of the gut microbiota–metabolism–immune axis, highlighting its potential as a non-invasive and cost-effective therapeutic approach for AD.
Full article
(This article belongs to the Special Issue Gut Microbiome in Homeostasis and Disease, 3rd Edition)
Open AccessReview
Evolution of Porcine Virus Isolation: Guidelines for Practical Laboratory Application
by
Danila Moiseenko, Roman Chernyshev, Natalya Kamalova, Vera Gavrilova and Alexey Igolkin
Microorganisms 2025, 13(12), 2658; https://doi.org/10.3390/microorganisms13122658 (registering DOI) - 22 Nov 2025
Abstract
Cell cultures are an essential tool for laboratory diagnosis of porcine viral infections. However, interpreting the results requires considering the species and tissue origin of cell lines as well as the specific virus replication characteristics (cytopathic effect). This guide discusses the development of
[...] Read more.
Cell cultures are an essential tool for laboratory diagnosis of porcine viral infections. However, interpreting the results requires considering the species and tissue origin of cell lines as well as the specific virus replication characteristics (cytopathic effect). This guide discusses the development of techniques for the primary isolation of viruses from biological material and provides recommendations for culturing viruses in different cell types. According to the World Organization for Animal Health, laboratory diagnosis should aim to isolate the virus in cell culture. We have studied the evolution of virus isolation methods for various diseases affecting pigs, including African swine fever virus (ASFV), classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (Aujeszky’s disease, PRV), rotaviruses (RV), teschoviruses (PTVs), swine pox virus (SwPV), swine influenza A virus (IAVs), parvovirus (PPV), coronaviruses, circoviruses (PCVs), diseases with vesicular syndrome, and others. During our analysis of the literature and our own experience, we found that the porcine kidney (PK-15) cell line is the most suitable for isolating most viral porcine pathogens. For ASFV and PRRSV, the porcine alveolar macrophages (PAMs) continue to remain the primary model for isolation. These findings can serve as a starting point for virological reference laboratories to select optimal conditions for cultivating, obtaining field isolates, and strain adaptation.
Full article
(This article belongs to the Section Virology)
Open AccessArticle
Biosynthesized Silver Selenide Nanoparticles from Meyerozyma guilliermondii as a Novel Adjuvant to Revolutionize Gentamicin Therapy
by
Min Xu, Lei Yang, Ya-Wei Zhang, Chao Wu, Yuan-Yuan Cheng and Hao Xue
Microorganisms 2025, 13(12), 2657; https://doi.org/10.3390/microorganisms13122657 (registering DOI) - 22 Nov 2025
Abstract
The increasing prevalence of antibiotic resistance necessitates the development of novel antimicrobial agents and therapeutic strategies. This study reports the extracellular biosynthesis of silver selenide nanoparticles (Ag2Se NPs) using Meyerozyma guilliermondii PG-1 and evaluates their antimicrobial and antibiofilm efficacy, both alone
[...] Read more.
The increasing prevalence of antibiotic resistance necessitates the development of novel antimicrobial agents and therapeutic strategies. This study reports the extracellular biosynthesis of silver selenide nanoparticles (Ag2Se NPs) using Meyerozyma guilliermondii PG-1 and evaluates their antimicrobial and antibiofilm efficacy, both alone and in combination with gentamicin. The NPs were thoroughly characterized, confirming their nanoscale size, crystallinity, and biomolecule-mediated stability. Ag2Se NPs exhibited broad-spectrum antibacterial activity against Gram-positive (Staphylococcus aureus) and Gram-negative (Pseudomonas aeruginosa, Escherichia coli) pathogens and showed strong synergy with gentamicin, particularly against P. aeruginosa and E. coli, as demonstrated through checkerboard and time–kill assays. The NPs also significantly inhibited biofilm formation and disrupted pre-formed biofilms. Mechanistic studies revealed that the antibacterial effects involved membrane disruption, ATP leakage, and elevated oxidative stress, while gene expression analysis in S. aureus indicated triggered stress responses related to biofilm formation. These findings suggest that biosynthesized Ag2Se NPs represent a promising synergistic agent for enhancing antibiotic efficacy and combating biofilm-related infections.
Full article
(This article belongs to the Topic Antimicrobial Agents and Nanomaterials—2nd Edition)
Open AccessArticle
Transfer and Fitness of ISAba52-Mediated tet(X3) Transposon in Acinetobacter spp.
by
Chong Chen, Jing Liu, Jie Gao, Yubing Hua, Taotao Wu and Jinlin Huang
Microorganisms 2025, 13(12), 2656; https://doi.org/10.3390/microorganisms13122656 (registering DOI) - 22 Nov 2025
Abstract
The global spread of tigecycline resistance conferred by tet(X3) poses a serious threat to clinical treatment of multidrug-resistant (MDR) Acinetobacter infections. Despite tet(X3) being detected in diverse Acinetobacter species, its transposition mechanism and fitness in these pathogens remain poorly characterized. Here,
[...] Read more.
The global spread of tigecycline resistance conferred by tet(X3) poses a serious threat to clinical treatment of multidrug-resistant (MDR) Acinetobacter infections. Despite tet(X3) being detected in diverse Acinetobacter species, its transposition mechanism and fitness in these pathogens remain poorly characterized. Here, we reported the first plasmid-borne ISAba52-mediated transposable element harboring tet(X3) in Acinetobacter amyesii YH16040. Conjugation experiments demonstrated the transferability of tet(X3) into the chromosome of Acinetobacter baylyi ADP1 at an efficiency of (7.1 ± 2.5) × 10−8. High-throughput sequencing revealed six tandem copies of ISAba52-flanked tet(X3), floR, and sul2 forming a 231.6 Kb complex transposon in the obtained transconjugant A. baylyi YH16040C. Phenotypic assays showed that YH16040C exhibited elevated resistance to tigecycline, chlortetracycline, florfenicol, and trimethoprim-sulfamethoxazole by 64- to 256-fold. Notably, YH16040C exhibited a growth advantage, reduced competition ability, and non-significant difference in biofilm formation compared to ADP1 in antibiotic-free backgrounds. Under moderate antibiotic treatment of tigecycline, chlortetracycline, florfenicol, and trimethoprim-sulfamethoxazole, the competition ability of YH16040C against ADP1 was significantly higher than that without antibiotics. All of these highlight the importance of ISAba52-mediated transposition in disseminating tet(X3) between Acinetobacter species and elucidate the fitness changes employed by MDR strains under antibiotic selection pressures. Our study advocates the urgent need for surveillance of ISAba52-associated resistance elements in human, animal, and environmental settings.
Full article
(This article belongs to the Section Antimicrobial Agents and Resistance)
►▼
Show Figures
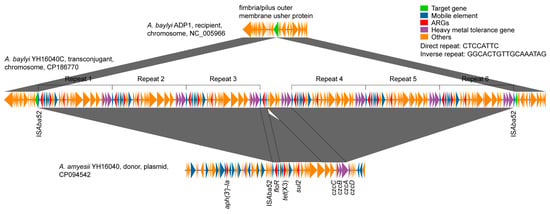
Figure 1
Open AccessArticle
Seven Years of Salmonella: Changing Resistance and Clinical Insights
by
Cristina Mihaela Sima, Aida Corina Bădescu, Georgiana Buruiană, Alexandru Duhaniuc, Luminița Smaranda Iancu, Eduard-Vasile Năstase and Olivia Simona Dorneanu
Microorganisms 2025, 13(12), 2655; https://doi.org/10.3390/microorganisms13122655 (registering DOI) - 22 Nov 2025
Abstract
Non-typhoidal Salmonella (NTS) represents a growing public health concern worldwide due to its increasing antimicrobial resistance and substantial disease burden, yet data from Romania remain limited. We conducted the first regional 7-year retrospective study of NTS-associated diarrhea among hospitalized patients, focusing on resistance
[...] Read more.
Non-typhoidal Salmonella (NTS) represents a growing public health concern worldwide due to its increasing antimicrobial resistance and substantial disease burden, yet data from Romania remain limited. We conducted the first regional 7-year retrospective study of NTS-associated diarrhea among hospitalized patients, focusing on resistance trends and clinical factors associated with disease severity. This study included all laboratory-confirmed Salmonella infections admitted for acute diarrheal disease to the “Sfânta Parascheva” Clinical Hospital of Infectious Diseases, Iași (January 2018–December 2024). Patient data were extracted from electronic medical records and analyzed using SPSS (v31.0), with statistical significance set at p < 0.05. Among the isolates obtained from the 698 included patients, most belonged to serogroup D (63.6%), followed by B (21.8%) and C (14.0%). Overall resistance rates were 12.3% for ampicillin, 3.6% for trimethoprim–sulfamethoxazole and 29.9% for ciprofloxacin, with a significant yearly increase observed only for ciprofloxacin (OR = 1.21, p < 0.001). Cardiovascular comorbidities were independently associated with prolonged hospitalization (>5 days) (OR = 2.25, p = 0.007). Invasive infections occurred in 14 patients (2%). Given the high ciprofloxacin resistance and the additional impact of comorbidities on disease severity, there is a need for ongoing surveillance and targeted management strategies.
Full article
(This article belongs to the Special Issue Infectious Disease Surveillance in Romania: Second Edition)
►▼
Show Figures
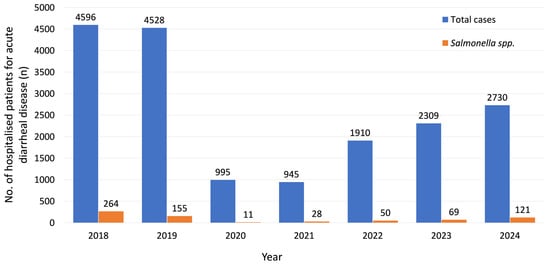
Figure 1
Open AccessArticle
Comprehensive Bioinformatics Analysis the circRNAs of Viral Infection Associated Pathway in HepG2 Expressing ORF3 of Genotype IV Swine Hepatitis E Virus
by
Hanwei Jiao, Lingjie Wang, Chi Meng, Shengping Wu, Yubo Qi, Jianhua Guo, Jixiang Li, Liting Cao, Yu Zhao, Jake J. Wen and Fengyang Wang
Microorganisms 2025, 13(12), 2654; https://doi.org/10.3390/microorganisms13122654 (registering DOI) - 22 Nov 2025
Abstract
The open reading frame 3 (ORF3) protein of the swine hepatitis E virus (SHEV) is a critical virulence factor implicated in viral infection, yet its precise mechanisms remain poorly understood. Circular RNAs (circRNAs) have emerged as key regulators of gene expression during viral
[...] Read more.
The open reading frame 3 (ORF3) protein of the swine hepatitis E virus (SHEV) is a critical virulence factor implicated in viral infection, yet its precise mechanisms remain poorly understood. Circular RNAs (circRNAs) have emerged as key regulators of gene expression during viral infections by functioning as miRNA sponges. This study aimed to identify key circRNAs and construct a potential circRNA-miRNA-mRNA regulatory network associated with the viral infection pathway in HepG2 cells expressing genotype IV SHEV ORF3. Based on our previous high-throughput circRNA and transcriptome sequencing data from HepG2 cells with adenovirus-mediated ORF3 overexpression, we screened for differentially expressed circRNAs and mRNAs linked to viral infection pathways. Using bioinformatic tools, we predicted miRNAs targeted by these mRNAs and those that could bind to the circRNAs, ultimately constructing a competing endogenous RNA (ceRNA) network with Cytoscape. We identified 31 differentially expressed circRNAs and 7 mRNAs (HSPA8, HSPA1B, EGR2, CXCR4, SOCS3, NOTCH3, and ZNF527) related to viral infection. A potential ceRNA network comprising 32 circRNAs, 23 miRNAs, and the 7 mRNAs was constructed. Core circRNAs, including ciRNA203, circRNA14936, and circRNA5562, may act as miRNA sponges to regulate the expression of these mRNAs. This network suggests a novel mechanism by which SHEV ORF3 might modulate host cell functions to facilitate viral infection.
Full article
(This article belongs to the Section Virology)
Open AccessArticle
Thuja sutchuenensis Franch. Essential Oil Ameliorates Atopic Dermatitis Symptoms in Mice by Modulating Skin Microbiota Composition and Reducing Inflammation
by
Nana Long, Youwei Zuo, Jian Li, Renxiu Yao, Quan Yang and Hongping Deng
Microorganisms 2025, 13(12), 2653; https://doi.org/10.3390/microorganisms13122653 (registering DOI) - 22 Nov 2025
Abstract
Atopic dermatitis (AD) is a chronic inflammatory skin disorder characterized by dysregulated immunity, skin barrier dysfunction, and cutaneous microbiome dysbiosis. While current therapies face limitations, Thuja sutchuenensis essential oil (TEO) shows promise due to its multi-target potential. We sought to explore the beneficial
[...] Read more.
Atopic dermatitis (AD) is a chronic inflammatory skin disorder characterized by dysregulated immunity, skin barrier dysfunction, and cutaneous microbiome dysbiosis. While current therapies face limitations, Thuja sutchuenensis essential oil (TEO) shows promise due to its multi-target potential. We sought to explore the beneficial effects of TEO and delve into its mechanistic actions in a mouse model of AD. We combined network pharmacology with in vivo validation to evaluate the therapeutic efficacy and mechanisms of TEO in an AD model, and confirmed network-predicted targets in an in vitro inflammatory cell model. In AD mice, TEO alleviated pruritus and epidermal hyperplasia, suppressed systemic IL-4/TNF-α and IgE, and partially normalized serum ALB, LDL-C, and HDL-C. Microbial diversity increased after treatment, although potentially pathogenic taxa (Arthrobacter sp. and Corynebacterium mastitidis) remained enriched. Machine-learning analysis indicated the highest predicted metabolic activity in CK controls, whereas the AD and TEO groups showed elevated pathogenic phenotype scores. Network pharmacology prioritized active compounds [(E)-ligustilide, senkyunolide A, 3-butylisobenzofuran-1(3H)-one, butylated hydroxytoluene, Z-buthlidenephthalide, and β-Myrcene] and core targets (TNF, PTPRC, CCR5, JAK1), implicating T-cell receptor signaling, Staphylococcus aureus infection, and STAT3 pathways. Docking and molecular dynamics supported strong, stable binding of major constituents to JAK1, and Western blotting confirmed TEO-mediated inhibition of the JAK1/STAT3 axis. TEO effectively alleviates atopic dermatitis symptoms by modulating immune responses and enhancing microbial diversity. It targets key signaling pathways, such as JAK1/STAT3, highlighting its potential as a therapeutic option for AD.
Full article
(This article belongs to the Section Medical Microbiology)
►▼
Show Figures

Figure 1
Open AccessArticle
Isolation and Characterization of a Duck Hepatitis A Virus Type 3 SH52 Strain in Ducklings
by
Minfan Huang, Dun Shuo, Yifei Xiong, Mei Tang, Yufei Wang, Xue Pan, Chunxiu Yuan, Qinfang Liu, Zhifei Zhang, Qiaoyang Teng, Bangfeng Xu, Xiaona Shi, Minghao Yan, Peirong Jiao, Zejun Li and Dawei Yan
Microorganisms 2025, 13(12), 2652; https://doi.org/10.3390/microorganisms13122652 (registering DOI) - 22 Nov 2025
Abstract
Duck hepatitis A virus type 3 (DHAV-3), an avian-specific pathogen primarily impacting ducklings, poses a significant threat to the duck farming industry by causing high mortality rates. A DHAV-3 strain SH52 was isolated from the diseased ducks and phylogenetic analysis of the whole
[...] Read more.
Duck hepatitis A virus type 3 (DHAV-3), an avian-specific pathogen primarily impacting ducklings, poses a significant threat to the duck farming industry by causing high mortality rates. A DHAV-3 strain SH52 was isolated from the diseased ducks and phylogenetic analysis of the whole genome revealed that the DHAV-3 SH52 belongs to the prevalent strains in China. The DHAV-3 SH52 strain replicates at high levels in various organs of 8-day-old specific pathogen-free (SPF) shelducks, causing pathological damage and leading to high lethality in 8-day-olds following intramuscular infection. The findings of this study provide an ideal animal challenge model for evaluating the efficacy of DHAV-3 vaccines in the future.
Full article
(This article belongs to the Special Issue Epidemiology, Detection and Control of Avian Infectious Diseases)
►▼
Show Figures
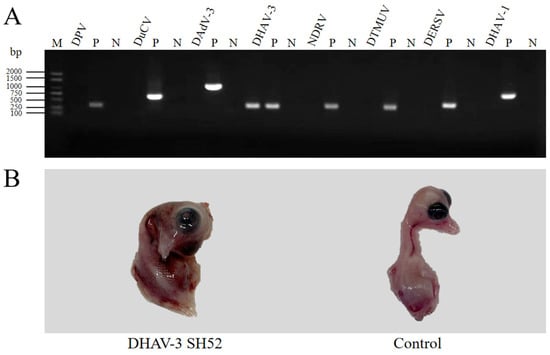
Figure 1
Open AccessArticle
Oral Administration of the Probiotic Lacticaseibacillus rhamnosus CA15 in a Large Cohort of Women with Bacterial Vaginosis and Mixed Vaginitis: Clinical Evidence from a Randomized, Double-Blind, Placebo-Controlled Study
by
Alessandra Pino, Amanda Vaccalluzzo, Stefano Cianci, Marco Palumbo, Giuseppe Caruso, Cinzia Caggia and Cinzia L Randazzo
Microorganisms 2025, 13(12), 2651; https://doi.org/10.3390/microorganisms13122651 - 21 Nov 2025
Abstract
Probiotics represent a valuable approach to boost vaginal health. A randomized double-blind placebo-controlled study was conducted to confirm the health benefits of the orally administered probiotic Lacticaseibacillus rhamnosus CA15 strain in a large cohort of women with bacterial vaginosis and mixed vaginitis, mainly
[...] Read more.
Probiotics represent a valuable approach to boost vaginal health. A randomized double-blind placebo-controlled study was conducted to confirm the health benefits of the orally administered probiotic Lacticaseibacillus rhamnosus CA15 strain in a large cohort of women with bacterial vaginosis and mixed vaginitis, mainly related to mild aerobic vaginitis and vulvovaginal candidiasis. Recruited women were randomly assigned in a 1:1 ratio to receive, for 10 days, oral capsules containing 1 × 1010 colony-forming units of the L. rhamnosus CA15 strain (Active group) or a placebo (Placebo group). Two-hundred women completed this study. Clinical and microbiological parameters were assessed at baseline (T0), 10 days (T1), and 30 days post-treatment (T2). In addition, quality of life was evaluated at T0 and T2. The study protocol was registered on clinicaltrials.gov (ID: NCT05796921). The oral administration of the L. rhamnosus CA15 strain resulted in significant improvements in clinical characteristics of vaginal dysbiosis as well as changes in the vaginal microbiota composition. Furthermore, participants allocated to the Active group reported enhancements in physical and psychological health, social relations, environment, and overall quality of life. No significant changes were observed in the Placebo group. The present study highlights the ability of the L. rhamnosus CA15 to manage vaginal dysbiosis, offering new perspectives for the treatment and prevention of common gynecological disorders.
Full article
(This article belongs to the Section Antimicrobial Agents and Resistance)
Open AccessArticle
Invertebrate-Derived DNA (iDNA) to Identify Sand Flies’ Bloodmeal: A Molecular Approach to Identifying Hosts in Blood-Feeding Vectors of Leishmaniasis
by
Bruno Oliveira Cova, Bruno Henrique Saranholi, Carla Cristina Gestich, Paulo Roberto Machado, Adriano Figueiredo Monte-Alegre and Albert Schriefer
Microorganisms 2025, 13(12), 2650; https://doi.org/10.3390/microorganisms13122650 - 21 Nov 2025
Abstract
DNA metabarcoding data obtained by next generation sequencing (NGS) has been used to identify species in mixed biological samples, such as DNA from the gut content of invertebrates that feed on vertebrates (invertebrate-derived DNA, iDNA). This investigation employed DNA metabarcoding approach to determine
[...] Read more.
DNA metabarcoding data obtained by next generation sequencing (NGS) has been used to identify species in mixed biological samples, such as DNA from the gut content of invertebrates that feed on vertebrates (invertebrate-derived DNA, iDNA). This investigation employed DNA metabarcoding approach to determine vertebrate hosts of female phlebotomine sand flies, blood-feeding leishmaniasis vectors. We evaluated performance across three mitochondrial markers: a mammal-specific mini-barcode (16S rRNA), a pan-vertebrate mini-barcode (12S rRNA), and a standard CytB barcode region. Phlebotomine sand flies collections occurred in the Cacao Region of Southeastern Bahia, Brazil, an American Tegumentary Leishmaniasis (ATL) endemic zone. Our analysis examined iDNA from forty female specimens pooled in thirteen samples of seven sand fly species, including confirmed ATL vectors. Metabarcoding-derived operational taxonomic units (OTUs) underwent taxonomic assignment through comparison with GenBank NCBI® reference databases. Results identified twenty vertebrate OTUs: primates (four OTUs), rodents (four), ungulates (five), marsupials (one), plus a domestic dog and a chicken. Notably, non-mammalian taxa, including reptiles (one OTU) and amphibians (three), were detected. The iDNA metabarcoding approach allowed us to accurately sample the diversity of phlebotomine sandflies’ bloodmeals in a single specimen of a non-engorged female sand fly with mixed feeding.
Full article
(This article belongs to the Special Issue Interactions between Parasites/Pathogens and Vectors)
Open AccessReview
Biofilm as a Key Element in the Bacterial Pathogenesis of Forest Trees: A Review of Mechanisms and Ecological Implications
by
Miłosz Tkaczyk
Microorganisms 2025, 13(12), 2649; https://doi.org/10.3390/microorganisms13122649 - 21 Nov 2025
Abstract
Bacterial diseases of forest trees represent an increasing threat to ecosystem health and the sustainability and resilience of forest management, particularly under changing climate conditions. One of the key yet still insufficiently understood adaptive mechanisms of pathogens is biofilm formation—a structured community of
[...] Read more.
Bacterial diseases of forest trees represent an increasing threat to ecosystem health and the sustainability and resilience of forest management, particularly under changing climate conditions. One of the key yet still insufficiently understood adaptive mechanisms of pathogens is biofilm formation—a structured community of bacterial cells embedded in a matrix of extracellular polymeric substances (EPS), which provides protection against stress factors, biocides, and the host’s defensive responses such as antimicrobial compounds or immune reactions.. This paper presents a comprehensive review of current knowledge on the role of biofilms in the bacterial pathogenesis of forest trees, covering their formation mechanisms, molecular regulation, and ecological significance. Four key stages of biofilm development are discussed—adhesion, microcolony formation, EPS production, and dispersion—along with the roles of quorum sensing systems and c-di-GMP-based signaling in regulating these processes. Examples of major tree pathogens are presented, including Pseudomonas syringae, Erwinia amylovora, Xylella fastidiosa, the Brenneria–Gibbsiella complex associated with Acute Oak Decline (AOD) andLonsdalea populi. Biofilm formation is shown to play a crucial role in the colonization of xylem, leaf surfaces, and tissues undergoing necrosis, where biofilms may stabilize decomposition zones and support saprophytic–pathogenic transitions. In the applied section, the concept of “biofilm-targeted control” is discussed, encompassing both chemical and biological strategies for disrupting biofilm structure—from quorum-sensing inhibitors and EPS-degrading enzymes to the use of biosurfactants and antagonistic microorganisms. The need for in situ research in forest environments and the adaptation of advanced imaging (CLSM, micro-CT) and metagenomic analyses to tree systems is also emphasized. This review concludes that biofilms are not merely a physiological form of bacterial organization but a complex adaptive system essential for the survival and virulence of pathogens in forest ecosystems. Understanding their functions is fundamental for developing sustainable and ecologically safe phytosanitary strategies for forest protection.
Full article
(This article belongs to the Special Issue Beneficial Biofilms: From Mechanisms to Applications)
Open AccessArticle
Effects of Human Trampling on Soil Microbial Community Assembly in Yangzhou Urban Forest Park
by
Jingwei Lian, Liwen Li, Xin Wan, Dongmei He, Yingzhou Tang, Wei Xing and Yingdan Yuan
Microorganisms 2025, 13(12), 2648; https://doi.org/10.3390/microorganisms13122648 - 21 Nov 2025
Abstract
Human trampling in urban forest parks has received increasing attention, yet its effects on microbial community assembly remain elusive. This study investigated how trampling influences soil physicochemical properties and microbial communities in Zhuyuwan Scenic Area. Neutral and null community models were used to
[...] Read more.
Human trampling in urban forest parks has received increasing attention, yet its effects on microbial community assembly remain elusive. This study investigated how trampling influences soil physicochemical properties and microbial communities in Zhuyuwan Scenic Area. Neutral and null community models were used to analyze the effects of trampling on microbial assembly processes. Trampling altered both soil physicochemical properties and microbial diversity. Fungal richness differed significantly between control and light-trampling plots. Soil bulk density (SD) was strongly negatively correlated with other soil physical properties, which were positively intercorrelated. Model analyses showed that light trampling strengthened stochastic processes in bacterial community assembly, whereas heavy trampling reduced this effect. Increasing trampling intensity intensified the influence of stochastic processes on fungal community assembly. Bacterial communities were mainly shaped by heterogeneous selection, while fungal communities were primarily governed by dispersal limitation. These results enhance understanding of how trampling disturbance influences microbial community assembly and provide a theoretical basis for the ecological management and restoration of urban forest parks.
Full article
(This article belongs to the Section Environmental Microbiology)
►▼
Show Figures

Figure 1
Open AccessArticle
Highly Alkaline-Resistant Enterococcus faecalis Induces Compromised M1 Polarization and Phagocytosis of Macrophage via Z-DNA Binding Protein 1
by
Yifang Xiao, Runze Liu, Yanling Yang, Xinlu Li, Yi Min and Wei Fan
Microorganisms 2025, 13(12), 2647; https://doi.org/10.3390/microorganisms13122647 - 21 Nov 2025
Abstract
Persistent apical periodontitis (PAP) of human teeth is related to Enterococcus faecalis (E. faecalis) with higher alkaline resistance. This study aimed to investigate how highly alkaline-resistant (HAR) E. faecalis modulates macrophage M1 polarization and phagocytosis via Z-DNA-binding protein 1 (ZBP1). HAR
[...] Read more.
Persistent apical periodontitis (PAP) of human teeth is related to Enterococcus faecalis (E. faecalis) with higher alkaline resistance. This study aimed to investigate how highly alkaline-resistant (HAR) E. faecalis modulates macrophage M1 polarization and phagocytosis via Z-DNA-binding protein 1 (ZBP1). HAR E. faecalis was generated through serial alkaline passaging. RAW264.7 macrophages were infected with standard or HAR E. faecalis. M1 polarization markers and Cd274 (Programmed death-ligand 1 (PD-L1)) were profiled by RT-qPCR. ZBP1 was detected by RT-qPCR and immunofluorescence staining, and was silenced using small interfering RNA (siRNA). iNOS, ZBP1 and PD-L1 proteins were analyzed by Western blotting. The phagocytosis of CFDA-SE-labeled bacteria was then quantified by confocal microscopy and flow cytometry. The results showed that HAR E. faecalis induced significantly lower expression of ZBP1, M1 polarization markers and Cd274 in macrophages than the standard strain. After ZBP1 knock-down, expression of these markers decreased. Macrophages phagocytosed much fewer HAR E. faecalis than the standard strain. After ZBP1 knock-down, the differences between the two strains disappeared. In conclusion, HAR E. faecalis induced compromised M1 polarization and phagocytosis of macrophage via ZBP1. These findings may provide new insights into the pathogenesis and treatment of PAP.
Full article
(This article belongs to the Section Molecular Microbiology and Immunology)
►▼
Show Figures

Figure 1
Open AccessArticle
Construction of Recombinant Escherichia coli Expressing Ammonia Assimilation Genes and Evaluation of Its Effect on Removing Ammonium Nitrogen (NH4+-N)
by
Pan Pan, Yongkun Yang, Runxuan Shi, Yulin Kang, Hanli Xu, Xiyu Cheng, Qiong Yan and Honggang Hu
Microorganisms 2025, 13(12), 2646; https://doi.org/10.3390/microorganisms13122646 - 21 Nov 2025
Abstract
Treating wastewater with high ammonium nitrogen (NH4+-N) is a public and environmental priority. Unlike nitrification–denitrification, ammonia assimilation channels NH4+-N into the glutamate biosynthetic pathway, avoiding gaseous nitrogen species (NOx, N2). Here we engineered Escherichia coli
[...] Read more.
Treating wastewater with high ammonium nitrogen (NH4+-N) is a public and environmental priority. Unlike nitrification–denitrification, ammonia assimilation channels NH4+-N into the glutamate biosynthetic pathway, avoiding gaseous nitrogen species (NOx, N2). Here we engineered Escherichia coli to enhance ammonia assimilation by co-expressing three key genes, gdhA, glnA, and guaA. The genes were synthesized, cloned into expression plasmids, and introduced into E. coli BL21 for IPTG-inducible expression. Expression of the target proteins at expected sizes was observed, and NH4+-N removal was assessed in flask fermentations. Recombinant strains exhibited significantly higher NH4+-N reduction than the empty vector control; among them, BL21(pET-gdhA-glnA-guaA) performed best, achieving a maximum removal efficiency of 90.09% under the tested conditions. These results indicate that reinforcing the glutamate pathway through multi-gene co-expression can effectively lower NH4+-N in culture and provide a basis for developing recombinant bacteria for practical sewage treatment.
Full article
(This article belongs to the Section Environmental Microbiology)
►▼
Show Figures

Figure 1
Open AccessReview
Biotic and Abiotic Factors on Rhizosphere Microorganisms in Grassland Ecosystems
by
Bademu Qiqige, Yuzhen Liu, Yu Tian, Li Liu, Weiwei Guo, Ping Wang, Dayou Zhou, Hui Wen, Qiuying Zhi, Yuxuan Wu, Xiaosheng Hu, Ming Li and Junsheng Li
Microorganisms 2025, 13(12), 2645; https://doi.org/10.3390/microorganisms13122645 - 21 Nov 2025
Abstract
Rhizosphere microbiota, serving as pivotal drivers of multifunctionality in grassland ecosystems, are jointly shaped by the dual influences of biotic and abiotic factors. Among biotic components, plant functional types selectively modulate microbial communities through root exudate specificity, while soil fauna (e.g., nematodes and
[...] Read more.
Rhizosphere microbiota, serving as pivotal drivers of multifunctionality in grassland ecosystems, are jointly shaped by the dual influences of biotic and abiotic factors. Among biotic components, plant functional types selectively modulate microbial communities through root exudate specificity, while soil fauna (e.g., nematodes and earthworms) drive microbial interaction networks via biophysical disturbances and trophic cascades. However, excessive nematode grazing suppresses the hyphal extension of arbuscular mycorrhizal fungi (AMF). Moderate grazing facilitates the proliferation of ammonia-oxidizing bacteria through fecal input, whereas intensive grazing induces topsoil compaction, leading to a dramatic 40–60% reduction in lipopolysaccharide content in Gram-negative bacteria. Long-term chemical fertilization significantly decreases the fungal-to-bacterial ratio, while organic amendments enhance microbial carbon use efficiency by activating extracellular enzymatic activities. Regarding abiotic factors, the stoichiometric characteristics of soil carbon, nitrogen, and phosphorus directly regulate microbial metabolic strategies. Hydrological dynamics influence microbial respiratory pathways through oxygen partial pressure shifts—drought stress inhibits mycelial network development. Future research should focus on predicting the emissions of gases such as N2O (ozone monomer) and optimizing nitrogen fertilizer management to significantly reduce greenhouse gas emissions at the source. The soil organic carbon storage in grassland ecosystems is extremely large. Effective prediction and management can make these soils become important carbon “sinks”, offsetting the carbon dioxide in the atmosphere. At the same time, transcriptomics and metabolic flux analysis should be combined with multi-omics technologies and in situ labeling methods to provide theoretical basis and technical support for developing mechanism-based and predictable grassland restoration and adaptive management strategies from both macroscopic and microscopic perspectives.
Full article
(This article belongs to the Section Environmental Microbiology)
►▼
Show Figures

Figure 1
Open AccessArticle
Downregulation of oar-miR-125b Drives Blood–Brain Barrier Breakdown Through the TNFSF4–NF-κB Inflammatory Axis in Enterococcus Faecalis Meningitis
by
Longling Jiao, You Wu, Borui Qi, Pengfei Zhao, Ming Zhou, Runze Zhang, Yongjian Li, Jingjing Ren, Shuzhu Cao and Yayin Qi
Microorganisms 2025, 13(12), 2644; https://doi.org/10.3390/microorganisms13122644 - 21 Nov 2025
Abstract
Bacterial meningitis involves complex molecular networks, including microRNA-mediated regulation of inflammatory responses; however, the specific role of Ovis aries microRNA-125b (oar-miR-125b) in this process remains poorly understood. In this study, using a lamb model of Enterococcus faecalis-induced meningitis, we observed significant downregulation of
[...] Read more.
Bacterial meningitis involves complex molecular networks, including microRNA-mediated regulation of inflammatory responses; however, the specific role of Ovis aries microRNA-125b (oar-miR-125b) in this process remains poorly understood. In this study, using a lamb model of Enterococcus faecalis-induced meningitis, we observed significant downregulation of oar-miR-125b, which inversely correlated with its newly identified target, Tumor Necrosis Factor Superfamily Member 4 (TNFSF4). Dual-luciferase reporter assays confirmed that oar-miR-125b directly binds to the 3′ Untranslated Region (3′UTR) of TNFSF4 but not to the 3′UTRs of Kelch Like Family Member 31 (KLHL31) or NF-κB Inhibitor Interacting Ras Like 2 (NKIRAS2). Mechanistically, decreased oar-miR-125b expression relieves its repression of TNFSF4, leading to NF-κB pathway activation and blood–brain barrier disruption. Collectively, our results demonstrate that oar-miR-125b serves as a key anti-inflammatory regulator in bacterial meningitis by targeting TNFSF4 and constraining NF-κB signaling, highlighting its potential as a therapeutic target for attenuating neuroinflammation in meningitis.
Full article
(This article belongs to the Section Molecular Microbiology and Immunology)
►▼
Show Figures

Figure 1
Open AccessArticle
Metagenomic Analysis of Gut Microbiota Structure and Function in Adults with Subclinical Hypothyroidism: A Cross-Sectional Study in China
by
Xueqing Li, Xue Ma, Lizhi Wu, Zhe Mo, Zhijian Chen, Ronghua Zhang and Mingluan Xing
Microorganisms 2025, 13(11), 2643; https://doi.org/10.3390/microorganisms13112643 - 20 Nov 2025
Abstract
Subclinical hypothyroidism (SCH) is a condition characterized by thyroid hormone dysregulation, often associated with subtle clinical symptoms and metabolic disturbances. Emerging evidence suggests that the gut microbiota plays a crucial role in modulating thyroid function, but the microbiota–thyroid axis in SCH remains poorly
[...] Read more.
Subclinical hypothyroidism (SCH) is a condition characterized by thyroid hormone dysregulation, often associated with subtle clinical symptoms and metabolic disturbances. Emerging evidence suggests that the gut microbiota plays a crucial role in modulating thyroid function, but the microbiota–thyroid axis in SCH remains poorly understood. This study systematically investigates the gut microbiota composition, functional characteristics, and their correlation with thyroid hormone profiles in SCH patients. Using metagenomic sequencing and thyroid function assessments, we identified significant alterations in the gut microbiota of SCH patients, including a depletion of beneficial microbes such as Blautia and Bifidobacterium, and an enrichment of opportunistic pathogens like Bacteroides and Escherichia. Notably, Blautia depletion was negatively correlated with TSH levels, while Bacteroides abundance positively correlated with TSH levels, further highlighting the role of gut microbiota in thyroid dysfunction. Moreover, functional gene analysis revealed significant alterations in microbial metabolic pathways, with key pathways demonstrating correlations with thyroid hormone levels (free triiodothyronine (FT3) and triiodothyronine (T3)). Our findings suggest that gut microbial dysbiosis is closely associated with SCH. The study provides novel insights into the gut–thyroid axis and its role in SCH, offering new targets for early diagnosis, risk stratification, and intervention strategies in thyroid diseases.
Full article
(This article belongs to the Section Public Health Microbiology)
►▼
Show Figures

Figure 1

Journal Menu
► ▼ Journal Menu-
- Microorganisms Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Biomass, Microorganisms, Sustainability, Water, Fermentation, Energies, Materials, Applied Biosciences
Recovery and Use of Bioactive Materials and Biomass
Topic Editors: Xiang Li, Tianfeng Wang, Xianbao XuDeadline: 25 November 2025
Topic in
Applied Microbiology, Bioengineering, Biology, Environments, Microorganisms
Environmental Bioengineering and Geomicrobiology
Topic Editors: Xian-Chun Zeng, Deng LiuDeadline: 20 December 2025
Topic in
Applied Microbiology, Fermentation, Foods, Microbiology Research, Microorganisms
Fermented Food: Health and Benefit
Topic Editors: Niel Van Wyk, Alice VilelaDeadline: 31 December 2025
Topic in
Applied Microbiology, Microbiology Research, Microorganisms, IJMS, IJPB, Plants
New Challenges on Plant–Microbe Interactions
Topic Editors: Wenfeng Chen, Junjie ZhangDeadline: 31 January 2026

Special Issues
Special Issue in
Microorganisms
Advances in Ochratoxin A Research—Implications for Detoxification, Food Safety, and Human Health
Guest Editor: José MancheñoDeadline: 30 November 2025
Special Issue in
Microorganisms
Diversity, Function, and Ecology of Soil Microbial Communities
Guest Editor: Adijailton José de SouzaDeadline: 30 November 2025
Special Issue in
Microorganisms
Microbial Gene Editing Technology
Guest Editors: Yeshi Yin, Aihua DengDeadline: 30 November 2025
Special Issue in
Microorganisms
Advances in Molecular Biology of Entamoeba histolyticaGuest Editor: Laurence A. MarchatDeadline: 30 November 2025
Topical Collections
Topical Collection in
Microorganisms
Device-Related Infections and Bacterial Biofilms
Collection Editor: María Guembe
Topical Collection in
Microorganisms
Feature Papers in Plant Microbe Interactions
Collection Editor: Rainer Borriss
Topical Collection in
Microorganisms
Feature Papers in Antimicrobial Agents and Resistance
Collection Editor: Maurizio Ciani
Topical Collection in
Microorganisms
Advances in SARS-CoV-2 Infection
Collection Editor: Carlo Contini








