Transfer and Fitness of ISAba52-Mediated tet(X3) Transposon in Acinetobacter spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Ethical Statement
2.2. Conjugation Experiments
2.3. Whole Genome Sequencing (WGS) and Bioinformatics Analyses
2.4. Antimicrobial Susceptibility Testing
2.5. Growth Curves
2.6. Competition Experiments
2.7. Crystal Violet Staining
2.8. Statistical Analyses
3. Results
3.1. Transferability of tet(X3)-Mediated Tigecycline Resistance
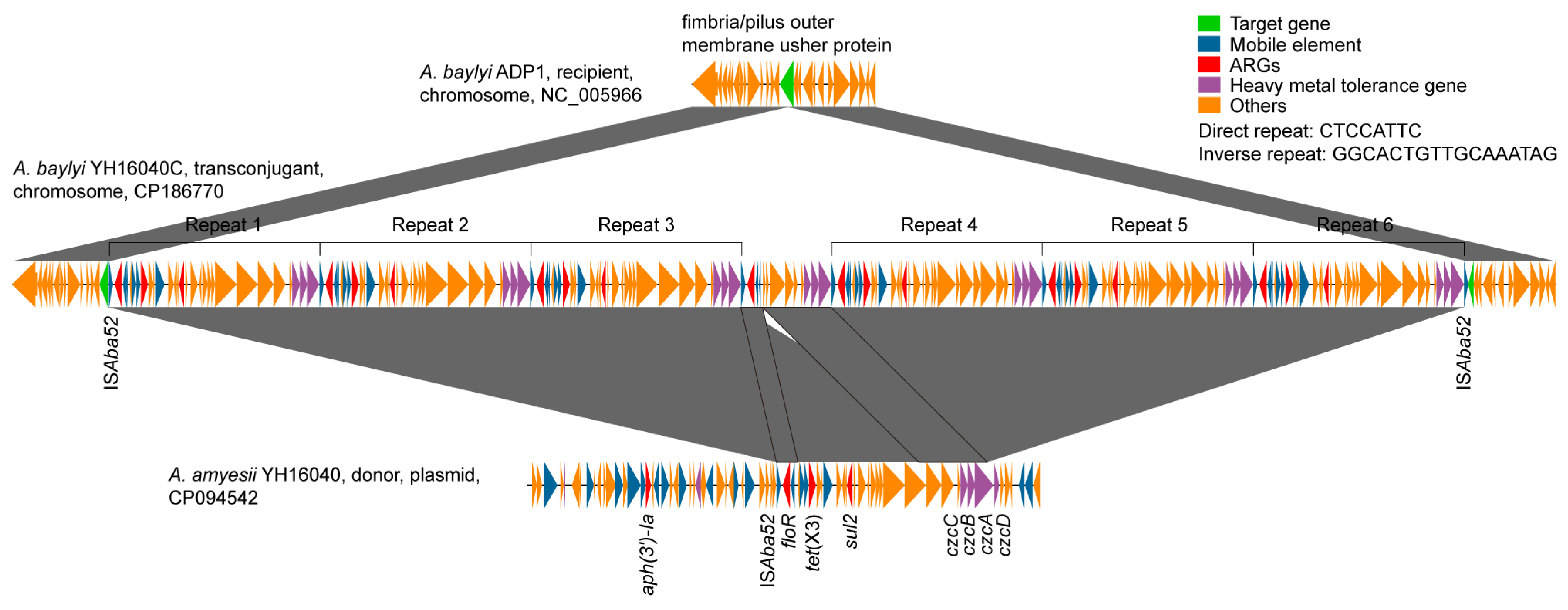
3.2. Transposition Mechanism of tet(X3) Across Acinetobacter Species
3.3. Fitness Effect of ISAba52-Mediated tet(X3) Transposon
3.3.1. Bacterial Growth
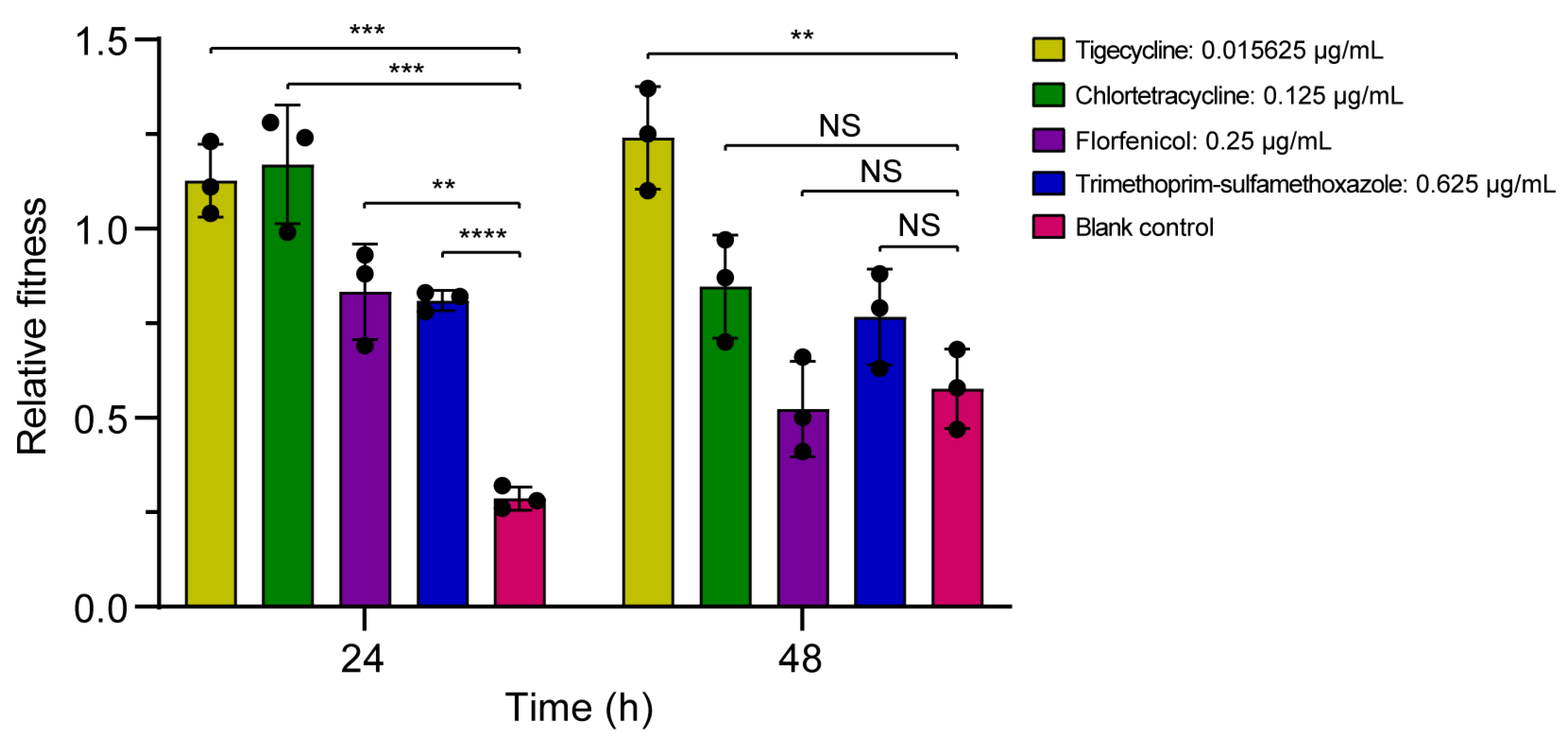
3.3.2. Bacterial Competition
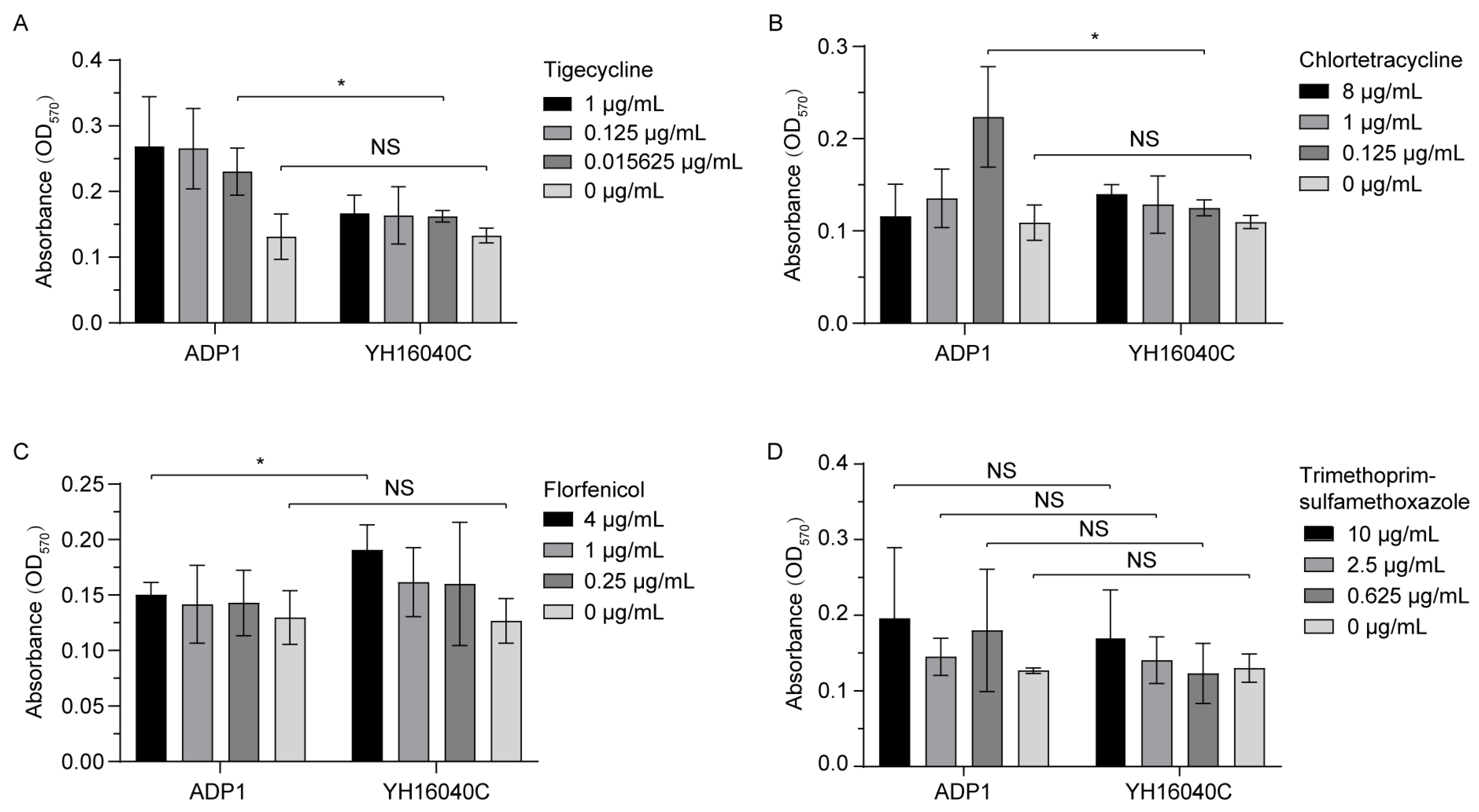
3.3.3. Bacterial Biofilm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MDR | Multidrug-resistant |
| FDA | Food and Drug Administration |
| CRAB | Carbapenem-resistant Acinetobacter baumannii |
| WHO | World Health Organization |
| ARGs | Antibiotic resistance genes |
| LB | Luria–Bertani |
| ERIC-PCR | Enterobacterial repetitive intergenic consensus PCR |
| SD | Standard deviation |
| WGS | Whole genome sequencing |
| RAST | Rapid annotation using subsystem technology |
| ORF | Open reading frame |
| NCBI | National Center for Biotechnology Information |
| ANI | Average nucleotide identity |
| CLSI | Clinical and Laboratory Standards Institute |
| MICs | Minimum inhibitory concentrations |
| MH | Mueller–Hinton |
| RF | Relative fitness |
| CFU | Colony-forming unit |
| PBS | Phosphate buffer saline |
| NS | Not significant |
| IS | Insertion sequence |
| RND | Resistance-nodulation-division |
References
- Seifert, H.; Blondeau, J.; Lucassen, K.; Utt, E.A. Global update on the in vitro activity of tigecycline and comparators against isolates of Acinetobacter baumannii and rates of resistant phenotypes (2016–2018). J. Glob. Antimicrob. Resist. 2022, 31, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-S.; Yang, J.-L.; Wang, J.-T.; Sheng, W.-H.; Yang, C.-J.; Chuang, Y.-C.; Chang, S.-C. Evaluation of the synergistic effect of eravacycline and tigecycline against carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae. J. Infect. Public Health 2024, 17, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; He, J.; Wang, J.; Zhang, L.; Zhang, L.; Xu, Q.; Shi, K.; Leptihn, S.; Shi, Y.; Fu, X.; et al. Novel tigecycline resistance mechanisms in Acinetobacter baumannii mediated by mutations in adeS, rpoB and rrf. Emerg. Microbes Infect. 2021, 10, 1404–1417. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Nwobi, O.C.; Okpala, C.O.R.; Ezeonu, I.M. Mobile Tigecycline Resistance: An Emerging Health Catastrophe Requiring Urgent One Health Global Intervention. Front. Microbiol. 2022, 13, 808744. [Google Scholar] [CrossRef]
- Shariati, A.; Dadashi, M.; Chegini, Z.; van Belkum, A.; Mirzaii, M.; Khoramrooz, S.S.; Darban-Sarokhalil, D. The global prevalence of Daptomycin, Tigecycline, Quinupristin/Dalfopristin, and Linezolid-resistant Staphylococcus aureus and coagulase–negative staphylococci strains: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control. 2020, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Wan, M.; Wang, C.; Gao, X.; Yang, Q.; Partridge, S.R.; Wang, Y.; Zong, Z.; Doi, Y.; Shen, J.; et al. Emergence of a Plasmid-Encoded Resistance-Nodulation-Division Efflux Pump Conferring Resistance to Multiple Drugs, Including Tigecycline, in Klebsiella pneumoniae. mBio 2020, 11, e02930-19. [Google Scholar] [CrossRef]
- Chiu, S.K.; Huang, L.Y.; Chen, H.; Tsai, Y.K.; Liou, C.H.; Lin, J.C.; Siu, L.K.; Chang, F.Y.; Yeh, K.M. Roles of ramR and tet(A) Mutations in Conferring Tigecycline Resistance in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates. Antimicrob. Agents Chemother. 2017, 61, e00391-17. [Google Scholar] [CrossRef]
- Liu, C.; Liu, J.; Lu, Q.; Wang, P.; Zou, Q. The Mechanism of Tigecycline Resistance in Acinetobacter baumannii under Sub-Minimal Inhibitory Concentrations of Tigecycline. Int. J. Mol. Sci. 2024, 25, 1819. [Google Scholar] [CrossRef]
- Yu, R.; Zhang, Y.; Xu, Y.; Schwarz, S.; Li, X.-S.; Shang, Y.-H.; Du, X.-D. Emergence of a tet(M) Variant Conferring Resistance to Tigecycline in Streptococcus suis. Front. Vet. Sci. 2021, 8, 709327. [Google Scholar] [CrossRef]
- Brajerova, M.; Nyc, O.; Drevinek, P.; Krutova, M. Genomic insights into the spread of vancomycin- and tigecycline-resistant Enterococcus faecium ST117. Ann. Clin. Microb. Antimicrob. 2025, 24, 36. [Google Scholar] [CrossRef]
- Yao, C.; Jin, L.; Wang, Q.; Wang, M.; Wang, R.; Cai, M.; Song, K.; Wang, H. Unraveling the evolution and global transmission of high level tigecycline resistance gene tet(X). Environ. Int. 2025, 199, 109499. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.J.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.B.; Ji, Q.J.; Wei, R.C.; Liu, Z.H.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wang, J.; Wang, Y.; Wang, L.; Bi, Y.; Zhu, B.; Gao, G.F. Tigecycline resistance tet(X3) gene is going wild. Biosaf. Health 2020, 2, 9–11. [Google Scholar] [CrossRef]
- Chen, C.; Cui, C.Y.; Yu, J.J.; He, Q.; Wu, X.T.; He, Y.Z.; Cui, Z.H.; Li, C.; Jia, Q.L.; Shen, X.G.; et al. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med. 2020, 12, 111. [Google Scholar] [CrossRef]
- Chen, C.; Wu, T.; Liu, J.; Gao, J. Threat and Control of tet(X)-Mediated Tigecycline-Resistant Acinetobacter sp. Bacteria. Foods 2025, 14, 3374. [Google Scholar] [CrossRef]
- Qin, J.; Feng, Y.; Lu, X.; Zong, Z. Precise Species Identification for Acinetobacter: A Genome-Based Study with Description of Two Novel Acinetobacter Species. mSystems 2021, 6, e00237-21. [Google Scholar] [CrossRef]
- Freese, H.M.; Meier-Kolthoff, J.P.; Sardà Carbasse, J.; O. Afolayan, A.; Göker, M. TYGS and LPSN in 2025: A Global Core Biodata Resource for genome-based classification and nomenclature of prokaryotes within DSMZ Digital Diversity. Nucleic Acids Res. 2025; Online ahead of print. [Google Scholar] [CrossRef]
- Al Atrouni, A.; Joly-Guillou, M.L.; Hamze, M.; Kempf, M. Reservoirs of Non-baumannii Acinetobacter Species. Front. Microbiol. 2016, 7, 49. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Luo, Q.; Chang, M.; Lu, P.; Guo, Q.; Jiang, X.; Xiao, T.; Zhang, H.; Ma, Y.; Zhang, Y.; Yu, W.; et al. Genomic epidemiology and phylodynamics of Acinetobacter baumannii bloodstream isolates in China. Nat. Commun. 2025, 16, 3536. [Google Scholar] [CrossRef] [PubMed]
- Mohd Sazlly Lim, S.; Zainal Abidin, A.; Liew, S.M.; Roberts, J.A.; Sime, F.B. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: A systematic review and meta-analysis. J. Infect. 2019, 79, 593–600. [Google Scholar] [CrossRef]
- WHO. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance. Available online: https://www.who.int/publications/i/item/9789240093461 (accessed on 30 September 2025).
- Cui, C.Y.; Chen, C.; Liu, B.T.; He, Q.; Wu, X.T.; Sun, R.Y.; Zhang, Y.; Cui, Z.H.; Guo, W.Y.; Jia, Q.L.; et al. Co-occurrence of Plasmid-Mediated Tigecycline and Carbapenem Resistance in Acinetobacter spp. from Waterfowls and Their Neighboring Environment. Antimicrob. Agents Chemother. 2020, 64, e02502-19. [Google Scholar] [CrossRef]
- Li, X.; Zhuang, Y.; Yu, Y.; Jia, H.; Kong, Y.; Zhang, J.; Xie, X.; Stehling, E.G.; Furlan, J.P.R.; Zhou, Z.; et al. Interplay of multiple carbapenemases and tigecycline resistance in Acinetobacter species: A serious combined threat. Clin. Microbiol. Infect. 2025, 31, 128–133. [Google Scholar] [CrossRef]
- Montaña, S.; Almuzara, M.; Pennini, M.; Sucari, A.; Centrón, D.; Vay, C.A.; Ramírez, M.S. ISCR2 and IS26: Two Insertion Sequences Highly Dispersed among Acinetobacter spp. Clinical Strains. J. Bacteriol. Mycol. Open Access 2017, 4, 33–36. [Google Scholar] [CrossRef]
- Yaffe, E.; Dethlefsen, L.; Patankar, A.V.; Gui, C.; Holmes, S.; Relman, D.A. Brief antibiotic use drives human gut bacteria towards low-cost resistance. Nature 2025, 641, 182–191. [Google Scholar] [CrossRef]
- Guo, Z.; Feng, S.; Liang, L.; Wu, Z.; Min, L.; Wang, R.; Li, J.; Zhong, L.-L.; Zhao, H.; Chen, X.; et al. Assessment of the reversibility of resistance in the absence of antibiotics and its relationship with the resistance gene’s fitness cost: A genetic study with mcr-1. Lancet Microbe 2024, 5, 100846. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.M.; Carfrae, L.A.; Rachwalski, K.; French, S.; Catacutan, D.; Gordzevich, R.; MacNair, C.R.; Speagle, M.E.; Werah, F.; Stokes, J.M.; et al. Exploiting the fitness cost of metallo-β-lactamase expression can overcome antibiotic resistance in bacterial pathogens. Nat. Microbiol. 2025, 10, 53–65. [Google Scholar] [CrossRef]
- Nemec, A.; Radolfová-Křížová, L.; Maixnerová, M.; Nemec, M.; Shestivska, V.; Španělová, P.; Kyselková, M.; Wilharm, G.; Higgins, P.G. Acinetobacter amyesii sp. nov., widespread in the soil and water environment and animals. Int. J. Syst. Evol. Microbiol. 2022, 72, 005642. [Google Scholar] [CrossRef] [PubMed]
- Dede, A.; Pérez-Valera, E.; Elhottová, D. Genome analysis of manure and soil-dwelling Acinetobacter strains indicates potential health risks associated with antibiotic resistance and virulence factors. Microb. Pathog. 2025, 205, 107610. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Clark, N.; Patel, J.B. pSK41-like plasmid is necessary for Inc18-like vanA plasmid transfer from Enterococcus faecalis to Staphylococcus aureus in vitro. Antimicrob. Agents Chemother. 2013, 57, 212–219. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Siguier, P. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- CLSI M100-Ed32; CLSI Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022.
- Ji, K.; Xu, Y.; Sun, J.; Huang, M.; Jia, X.; Jiang, C.; Feng, Y. Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 2019, 11, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Yi, L.X.; Yu, L.F.; Wang, J.; Liu, Y.; Chen, X.; Lv, L.; Yang, J.; Liu, J.H. Fitness Advantage of mcr-1-Bearing IncI2 and IncX4 Plasmids in Vitro. Front. Microbiol. 2018, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Machuca, J.; Briales, A.; Labrador, G.; Diaz-de-Alba, P.; Lopez-Rojas, R.; Docobo-Perez, F.; Martinez-Martinez, L.; Rodriguez-Bano, J.; Pachon, M.E.; Pascual, A.; et al. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J. Antimicrob. Chemother. 2014, 69, 3203–3215. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, P.-Y.; Cui, C.-Y.; He, Q.; Sun, J.; Liu, Y.-H.; Huang, J.-L. Classification and molecular characteristics of tet(X)-carrying plasmids in Acinetobacter species. Front. Microbiol. 2022, 13, 974432. [Google Scholar] [CrossRef]
- Zhang, R.-M.; Sun, J.; Sun, R.-Y.; Wang, M.-G.; Cui, C.-Y.; Fang, L.-X.; Liao, M.-N.; Lu, X.-Q.; Liu, Y.-X.; Liao, X.-P.; et al. Source Tracking and Global Distribution of the Tigecycline Non-Susceptible tet(X). Microbiol. Spectr. 2021, 9, e01164-21, Erratum in Microbiol. Spectr. 2022, 10, e01131-22. https://doi.org/10.1128/spectrum.01131-22. [Google Scholar] [CrossRef]
- Cheng, Y.Y.; Liu, Y.; Chen, Y.; Huang, F.M.; Chen, R.C.; Xiao, Y.H.; Zhou, K. Sporadic Dissemination of tet(X3) and tet(X6) Mediated by Highly Diverse Plasmidomes among Livestock-Associated Acinetobacter. Microbiol. Spectr. 2021, 9, e01141-21. [Google Scholar] [CrossRef]
- Cheng, Y.M.; Li, Y.K.; Yu, R.H.; Ma, M.X.; Yang, M.; Si, H.B. Identification of Novel tet(X3) Variants Resistant To Tigecycline in Acinetobacter Species. Microbiol. Spectr. 2022, 10, e01333-22. [Google Scholar] [CrossRef]
- Wang, J.; Lu, M.J.; Wu, H.; Wang, Z.Y.; Mei, C.Y.; Tian, Y.Q.; Pan, Z.M.; Jiao, X. Plasmid-borne tet(X3) and chromosome-borne tet(X6) in porcine Acinetobacter isolates. J. Glob. Antimicrob. Resist. 2022, 29, 17–19. [Google Scholar] [CrossRef]
- Wei, D.-W.; Wong, N.-K.; Song, Y.; Zhang, G.; Wang, C.; Li, J.; Feng, J.; Bonomo, R.A. IS26 Veers Genomic Plasticity and Genetic Rearrangement toward Carbapenem Hyperresistance under Sublethal Antibiotics. mBio 2022, 13, e03340-21, Erratum in mBio 2022, 13, e00414-22. https://doi.org/10.1128/mbio.00414-22. [Google Scholar] [CrossRef]
- Girgis, H.S.; DuPai, C.D.; Lund, J.; Reeder, J.; Guillory, J.; Durinck, S.; Liang, Y.; Kaminker, J.; Smith, P.A.; Skippington, E. Single-molecule nanopore sequencing reveals extreme target copy number heterogeneity in arylomycin-resistant mutants. Proc. Natl. Acad. Sci. USA 2020, 118, e2021958118. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Andersson, D.I. Bacteria can compensate the fitness costs of amplified resistance genes via a bypass mechanism. Nat. Commun. 2024, 15, 2333. [Google Scholar] [CrossRef]
- Fuzi, M. The fitness connection of antibiotic resistance. Front. Microbiol. 2025, 16, 1556656. [Google Scholar] [CrossRef]
- Jo, J.; Kim, S.J.; Kwon, K.T.; Ko, K.S. Resilience of tigecycline heteroresistance phenotype in Acinetobacter baumannii. J. Antimicrob. Chemother. 2025, 80, 496–502. [Google Scholar] [CrossRef]
- Jiang, L.; Cai, W.; Tang, F.; Wang, Z.; Liu, Y. Characterization of Fitness Cost Caused by Tigecycline-Resistance Gene tet(X6) in Different Host Bacteria. Antibiotics 2021, 10, 1172. [Google Scholar] [CrossRef]
- Zhu, M.; Dai, X. Shaping of microbial phenotypes by trade-offs. Nat. Commun. 2024, 15, 4238. [Google Scholar] [CrossRef]
- Heilbronner, S.; Krismer, B.; Brötz-Oesterhelt, H.; Peschel, A. The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol. 2021, 19, 726–739. [Google Scholar] [CrossRef]
- Batsch, M.; Guex, I.; Todorov, H.; Heiman, C.M.; Vacheron, J.; Vorholt, J.A.; Keel, C.; van der Meer, J.R. Fragmented micro-growth habitats present opportunities for alternative competitive outcomes. Nat. Commun. 2024, 15, 7591. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Wu, Y.; Li, S.; Hu, J.; Sun, W.; Ni, J. Profiles, drivers, and prioritization of antibiotics in China’s major rivers. J. Hazard. Mater. 2024, 477, 135399. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Duan, Y.J.; Wang, S.P.; Wang, L.T.; Hou, Z.L.; Cui, Y.X.; Hou, J.; Das, R.; Mao, D.Q.; Luo, Y. Occurrence and distribution of clinical and veterinary antibiotics in the faeces of a Chinese population. J. Hazard. Mater. 2019, 383, 121129. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, Y.; Liu, D.; Yang, D.; Liu, Z.; Wang, Y.; Wang, J.; Wang, X.; Xu, X.; Li, X.; et al. Abundance of tigecycline resistance genes and association with antibiotic residues in Chinese livestock farms. J. Hazard. Mater. 2021, 409, 124921. [Google Scholar] [CrossRef]
- Wan, Y.P.; Liu, Z.H.; Liu, Y. Veterinary antibiotics in swine and cattle wastewaters of China and the United States: Features and differences. Water Environ. Res. 2021, 93, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Blake, K.S.; Xue, Y.-P.; Gillespie, V.J.; Fishbein, S.R.S.; Tolia, N.H.; Wencewicz, T.A.; Dantas, G. The tetracycline resistome is shaped by selection for specific resistance mechanisms by each antibiotic generation. Nat. Commun. 2025, 16, 1452. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Impalli, I.; Poleon, S.; Denoel, P.; Cipriano, M.; Van Boeckel, T.P.; Pecetta, S.; Bloom, D.E.; Nandi, A. Global trends in antibiotic consumption during 2016–2023 and future projections through 2030. Proc. Natl. Acad. Sci. USA 2024, 121, e2411919121. [Google Scholar] [CrossRef] [PubMed]
- ECDC; EFSA; EMA. ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals. EFSA J. 2017, 15, e04872. [Google Scholar] [CrossRef]
- Anedda, E.; Farrell, M.L.; Morris, D.; Burgess, C.M. Evaluating the impact of heavy metals on antimicrobial resistance in the primary food production environment: A scoping review. Environ. Pollut. 2023, 320, 121035. [Google Scholar] [CrossRef]
- Murray, L.M.; Hayes, A.; Snape, J.; Kasprzyk-Hordern, B.; Gaze, W.H.; Murray, A.K. Co-selection for antibiotic resistance by environmental contaminants. NPJ Antimicrob. Resist. 2024, 2, 9. [Google Scholar] [CrossRef] [PubMed]

| Strains | Tigecycline | Chlortetracycline | Florfenicol | Trimethoprim-Sulfamethoxazole a | CuSO4 | CdCl2 |
|---|---|---|---|---|---|---|
| A. amyesii YH16040 | 8 | 64 | 32 | 160 | 500 | 12.5 |
| A. baylyi YH16040C | 8 | 64 | 64 | 160 | 500 | 12.5 |
| A. baylyi ADP1 | 0.03125 | 0.25 | 1 | 2.5 | 500 | 12.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Liu, J.; Gao, J.; Hua, Y.; Wu, T.; Huang, J. Transfer and Fitness of ISAba52-Mediated tet(X3) Transposon in Acinetobacter spp. Microorganisms 2025, 13, 2656. https://doi.org/10.3390/microorganisms13122656
Chen C, Liu J, Gao J, Hua Y, Wu T, Huang J. Transfer and Fitness of ISAba52-Mediated tet(X3) Transposon in Acinetobacter spp. Microorganisms. 2025; 13(12):2656. https://doi.org/10.3390/microorganisms13122656
Chicago/Turabian StyleChen, Chong, Jing Liu, Jie Gao, Yubing Hua, Taotao Wu, and Jinlin Huang. 2025. "Transfer and Fitness of ISAba52-Mediated tet(X3) Transposon in Acinetobacter spp." Microorganisms 13, no. 12: 2656. https://doi.org/10.3390/microorganisms13122656
APA StyleChen, C., Liu, J., Gao, J., Hua, Y., Wu, T., & Huang, J. (2025). Transfer and Fitness of ISAba52-Mediated tet(X3) Transposon in Acinetobacter spp. Microorganisms, 13(12), 2656. https://doi.org/10.3390/microorganisms13122656






