Seven Years of Salmonella: Changing Resistance and Clinical Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Data Collection
2.2. Statistical Analysis
2.3. Ethics Statement
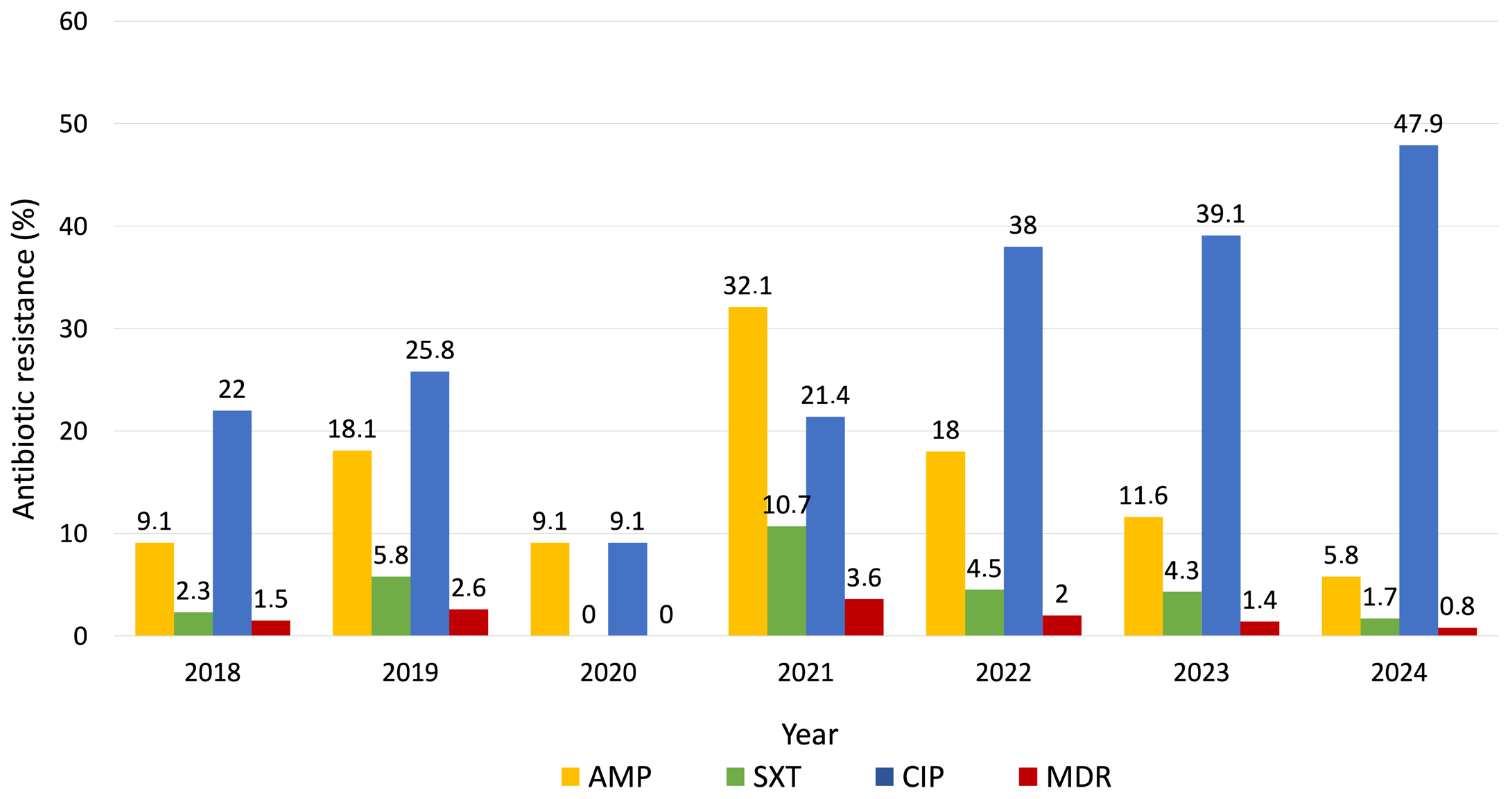
3. Results
3.1. Baseline Profile of the Study Population
3.2. Characterization of Circulating Salmonella Serogroups
3.3. Risk Factors for Prolonged Hospitalization
3.4. Invasive Salmonella Infections
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NTS | Non-typhoidal Salmonella |
| iNTS | Invasive non-typhoidal Salmonella |
| WBC | White blood cell |
| RBC | Red blood cell |
| CRP | C-Reactive Protein |
| AMP | Ampicillin |
| SXT | Trimethoprim-sulfamethoxazole |
| CIP | Ciprofloxacin |
| MDR | Multidrug resistance |
| EFSA | European Food Safety Authority |
| ECDC | European Centre for Disease Prevention and Control |
References
- Antony, B. Non Typhoidal Salmonellae and Its Aetiological Spectrum—An Overview with Indian Perspective. IP Int. J. Med. Microbiol. Trop. Dis. 2022, 8, 3–9. [Google Scholar] [CrossRef]
- Mulla, Z.D. Re: “Epidemiology of Salmonellosis in California, 1990–1999: Morbidity, Mortality, and Hospitalization Costs”. Am. J. Epidemiol. 2004, 159, 104. [Google Scholar] [CrossRef] [PubMed]
- Sanni, A.O.; Onyango, J.; Rota, A.F.; Mikecz, O.; Usman, A.; PicaCiamarra, U.; Fasina, F.O. Underestimated Economic and Social Burdens of Non-Typhoidal Salmonella Infections: The One Health Perspective from Nigeria. One Health 2023, 16, 100546. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, H.; Excler, J.-L.; Kim, J.H.; Lee, J.-S. The Economic Burden of Non-Typhoidal Salmonella and Invasive Non-Typhoidal Salmonella Infection: A Systematic Literature Review. Vaccines 2024, 12, 758. [Google Scholar] [CrossRef]
- Nemhauser, J.B.; Centers for Disease Control (U.S.) (Eds.) CDC Yellow Book 2024: Health Information for International Travel; Oxford University Press: New York, NY, USA, 2023. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2021–2022. EFSA J. 2024, 22, e8583. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Havelaar, A.H.; Ivarsson, S.; Löfdahl, M.; Nauta, M.J. Estimating the True Incidence of Campylobacteriosis and Salmonellosis in the European Union, 2009. Epidemiol. Infect. 2013, 141, 293–302. [Google Scholar] [CrossRef]
- O’Boyle, H.; Kirpalani, A.; Weiss, L.; Hames, N.; Li, R.; Leong, T.; Gonzalez, M.; Shane, A.L.; Charvat, C. Management and Outcomes of Salmonella Gastroenteritis in the Era of Rapid Molecular Testing. Hosp. Pediatr. 2022, 12, 1011–1019. [Google Scholar] [CrossRef]
- Galán-Relaño, Á.; Valero Díaz, A.; Huerta Lorenzo, B.; Gómez-Gascón, L.; Mena Rodríguez, M.Á.; Carrasco Jiménez, E.; Pérez Rodríguez, F.; Astorga Márquez, R.J. Salmonella and Salmonellosis: An Update on Public Health Implications and Control Strategies. Animals 2023, 13, 3666. [Google Scholar] [CrossRef]
- Dudhane, R.A.; Bankar, N.J.; Shelke, Y.P.; Badge, A.K. The Rise of Non-Typhoidal Salmonella Infections in India: Causes, Symptoms, and Prevention. Cureus 2023, 15, e46699. [Google Scholar] [CrossRef]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, Food Safety and Food Handling Practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef] [PubMed]
- Uzairue, L.I.; Shittu, O.B.; Ojo, O.E.; Obuotor, T.M.; Olanipekun, G.; Ajose, T.; Arogbonlo, R.; Medugu, N.; Ebruke, B.; Obaro, S.K. Antimicrobial Resistance and Virulence Genes of Invasive Salmonella enterica from Children with Bacteremia in North-Central Nigeria. SAGE Open Med. 2023, 11, 205031212311753. [Google Scholar] [CrossRef] [PubMed]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef]
- Chen, H.; Qiu, H.; Zhong, H.; Cheng, F.; Wu, Z.; Shi, T. Non-Typhoidal Salmonella Infections Among Children in Fuzhou, Fujian, China: A 10-Year Retrospective Review from 2012 to 2021. Infect. Drug Resist. 2023, 16, 2737–2749. [Google Scholar] [CrossRef] [PubMed]
- Stanaway, J.D.; Parisi, A.; Sarkar, K.; Blacker, B.F.; Reiner, R.C.; Hay, S.I.; Nixon, M.R.; Dolecek, C.; James, S.L.; Mokdad, A.H.; et al. The Global Burden of Non-Typhoidal Salmonella Invasive Disease: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Marchello, C.S.; Birkhold, M.; Crump, J.A.; Martin, L.B.; Ansah, M.O.; Breghi, G.; Canals, R.; Fiorino, F.; Gordon, M.A.; Kim, J.-H.; et al. Complications and Mortality of Non-Typhoidal Salmonella Invasive Disease: A Global Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2022, 22, 692–705. [Google Scholar] [CrossRef]
- Uche, I.V.; MacLennan, C.A.; Saul, A. A Systematic Review of the Incidence, Risk Factors and Case Fatality Rates of Invasive Nontyphoidal Salmonella (iNTS) Disease in Africa (1966 to 2014). PLoS Negl. Trop. Dis. 2017, 11, e0005118. [Google Scholar] [CrossRef]
- Morpeth, S.C.; Ramadhani, H.O.; Crump, J.A. Invasive Non-Typhi Salmonella Disease in Africa. Clin. Infect. Dis. 2009, 49, 606–611. [Google Scholar] [CrossRef]
- Zha, L.; Garrett, S.; Sun, J. Salmonella Infection in Chronic Inflammation and Gastrointestinal Cancer. Diseases 2019, 7, 28. [Google Scholar] [CrossRef]
- Van Elsland, D.M.; Duijster, J.W.; Zhang, J.; Stévenin, V.; Zhang, Y.; Zha, L.; Xia, Y.; Franz, E.; Sun, J.; Mughini-Gras, L.; et al. Repetitive Non-Typhoidal Salmonella Exposure Is an Environmental Risk Factor for Colon Cancer and Tumor Growth. Cell Rep. Med. 2022, 3, 100852. [Google Scholar] [CrossRef]
- Lamichhane, B.; Mawad, A.M.M.; Saleh, M.; Kelley, W.G.; Harrington, P.J.; Lovestad, C.W.; Amezcua, J.; Sarhan, M.M.; El Zowalaty, M.E.; Ramadan, H.; et al. Salmonellosis: An Overview of Epidemiology, Pathogenesis, and Innovative Approaches to Mitigate the Antimicrobial Resistant Infections. Antibiotics 2024, 13, 76. [Google Scholar] [CrossRef]
- Eikmeier, D.; Medus, C.; Smith, K. Incubation Period for Outbreak-Associated, Non-Typhoidal Salmonellosis Cases, Minnesota, 2000–2015. Epidemiol. Infect. 2018, 146, 423–429. [Google Scholar] [CrossRef]
- Motladiile, T.W.; Tumbo, J.M.; Malumba, A.; Adeoti, B.; Masekwane, N.J.; Mokate, O.M.R.; Sebekedi, O.C. Salmonella Food-Poisoning Outbreak Linked to the National School Nutrition Programme, North West Province, South Africa. S. Afr. J. Infect. Dis. 2019, 34, 124. [Google Scholar] [CrossRef]
- Yang, X.; Jin, K.; Yang, F.; Yuan, G.; Liu, W.; Xiang, L.; Wu, Z.; Li, Z.; Mao, J.; Shen, J.; et al. Nontyphoidal Salmonella Gastroenteritis in Baoshan, Shanghai, China, 2010 to 2014: An Etiological Surveillance and Case-Control Study. J. Food Prot. 2017, 80, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Sevilla-Navarro, S.; Catalá-Gregori, P.; García, C.; Cortés, V.; Marin, C. Salmonella Infantis and Salmonella Enteritidis Specific Bacteriophages Isolated Form Poultry Faeces as a Complementary Tool for Cleaning and Disinfection against Salmonella. Comp. Immunol. Microbiol. Infect. Dis. 2020, 68, 101405. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-J.; Chen, Y.-C.; Chen, N.-W.; Hsu, Y.-J.; Chu, H.-H.; Chen, C.-L.; Chiu, C.-H. Changing Antimicrobial Resistance and Epidemiology of Non-Typhoidal Salmonella Infection in Taiwanese Children. Front. Microbiol. 2021, 12, 648008. [Google Scholar] [CrossRef]
- Hengkrawit, K.; Tangjade, C. Prevalence and Trends in Antimicrobial Susceptibility Patterns of Multi-Drug-Resistance Non-Typhoidal Salmonella in Central Thailand, 2012–2019. Infect. Drug Resist. 2022, 15, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Sarkodie-Addo, P.; Aglomasa, B.C.; Donkor, E.S. Prevalence and Antimicrobial Resistance Patterns of Nontyphoidal Salmonella in Ghana: A Systematic Review and Meta-Analysis. Trop. Med. Health 2025, 53, 91. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Han, H.; Liu, J.; Ke, B.; Zhan, L.; Yang, X.; Tan, D.; Yu, B.; Huo, X.; Ma, X.; et al. Antimicrobial Resistance Profiles of Salmonella Isolates from Human Diarrhea Cases in China: An Eight-Year Surveilance Study. One Health Adv. 2023, 1, 2. [Google Scholar] [CrossRef]
- Gong, B.; Li, H.; Feng, Y.; Zeng, S.; Zhuo, Z.; Luo, J.; Chen, X.; Li, X. Prevalence, Serotype Distribution and Antimicrobial Resistance of Non-Typhoidal Salmonella in Hospitalized Patients in Conghua District of Guangzhou, China. Front. Cell. Infect. Microbiol. 2022, 12, 805384. [Google Scholar] [CrossRef]
- Crump, J.A.; Nyirenda, T.S.; Kalonji, L.M.; Phoba, M.-F.; Tack, B.; Platts-Mills, J.A.; Gordon, M.A.; Kariuki, S.M. Nontyphoidal Salmonella Invasive Disease: Challenges and Solutions. Open Forum Infect. Dis. 2023, 10 (Suppl. S1), S32–S37. [Google Scholar] [CrossRef]
- Tăbăran, A.; Dan, S.D.; Colobaţiu, L.M.; Mihaiu, M.; Condor, S.; Mărgăoan, R.; Crişan-Reget, O.L. Evaluation of Multidrug Resistance of Salmonella Isolated from Pork Meat Obtained from Traditional Slaughter Systems in Romania. Microorganisms 2024, 12, 2196. [Google Scholar] [CrossRef]
- Forgaciu, A.; Tabaran, A.; Colobatiu, L.; Mihaiu, R.; Dan, S.D.; Mihaiu, M. Concerning Increase in Antimicrobial Resistance Patterns of Pathogenic Strains of Salmonella Isolated in Poultry Meat Products. Antibiotics 2022, 11, 1469. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, W.; Zhang, R.; Wen, J.; Liu, S.; Jiang, Y.; Lin, L.; Chen, W.; Liang, J.; Ma, X.; et al. Epidemiological Features of Nontyphoidal Salmonella Infections Reported to Foodborne Disease Surveillance System in China, 2013–2022. BMC Public Health 2025, 25, 2258. [Google Scholar] [CrossRef]
- Assefa, A.; Girma, M. Prevalence and Antimicrobial Susceptibility Patterns of Salmonella and Shigella Isolates among Children Aged below Five Years with Diarrhea Attending Robe General Hospital and Goba Referral Hospital, South East Ethiopia. Trop. Dis. Travel Med. Vaccines 2019, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Frost, I.; Naylor, N.R.; Au, H.; Kim, Y.; Lee, Y.; Bzymek, A.; Majgier, K.; Moldoveanu, A.L.; Salman, O.M.; et al. Length of Hospital Stay and Associated Treatment Costs for Patients with Susceptible and Antibiotic-Resistant Salmonella Infections: A Systematic Review and Meta-Analysis. BMJ Open 2025, 15, e092494. [Google Scholar] [CrossRef] [PubMed]
- Gil Prieto, R.; Alejandre, C.G.; Meca, A.Á.; Barrera, V.H.; De Miguel, Á.G. Epidemiology of Hospital-Treated Salmonella Infection; Data from a National Cohort over a Ten-Year Period. J. Infect. 2009, 58, 175–181. [Google Scholar] [CrossRef]
- Hagedoorn, N.N.; Murthy, S.; Birkhold, M.; Marchello, C.S.; Crump, J.A.; The Vacc-iNTS Consortium Collaborators. Prevalence and Distribution of Non-Typhoidal Salmonella Enterica Serogroups and Serovars Isolated from Normally Sterile Sites: A Global Systematic Review. Epidemiol. Infect. 2024, 152, e4. [Google Scholar] [CrossRef]
- Kumar, G.; Kumar, S.; Jangid, H.; Dutta, J.; Shidiki, A. The Rise of Non-Typhoidal Salmonella: An Emerging Global Public Health Concern. Front. Microbiol. 2025, 16, 1524287. [Google Scholar] [CrossRef]
- Bloomfield, S.J.; Janecko, N.; Palau, R.; Alikhan, N.-F.; Mather, A.E. Genomic Diversity and Epidemiological Significance of Non-Typhoidal Salmonella Found in Retail Food Collected in Norfolk, UK. Microb. Genomics 2023, 9, 001075. [Google Scholar] [CrossRef]
- Mkangara, M. Prevention and Control of Human Salmonella Enterica Infections: An Implication in Food Safety. Int. J. Food Sci. 2023, 2023, 8899596. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- Alvarez, D.M.; Barrón-Montenegro, R.; Conejeros, J.; Rivera, D.; Undurraga, E.A.; Moreno-Switt, A.I. A Review of the Global Emergence of Multidrug-Resistant Salmonella Enterica Subsp. Enterica Serovar Infantis. Int. J. Food Microbiol. 2023, 403, 110297. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.-B.; Song, B.-J.; Shin, M.-Y.; Lim, H.-C.; Yoon, Y.-H.; Jeon, D.-Y.; Ha, H.; Yang, S.-I.; Kim, J.-B. Antibiotic Resistance Patterns and Serotypes of Salmonella Spp. Isolated at Jeollanam-Do in Korea. Osong Public Health Res. Perspect. 2017, 8, 211–219. [Google Scholar] [CrossRef]
- Herbinger, K.-H.; Hanus, I.; Schunk, M.; Beissner, M.; Von Sonnenburg, F.; Löscher, T.; Bretzel, G.; Hoelscher, M.; Nothdurft, H.D.; Huber, K.L. Elevated Values of C-Reactive Protein Induced by Imported Infectious Diseases: A Controlled Cross-Sectional Study of 11,079 Diseased German Travelers Returning from the Tropics and Subtropics. Am. Soc. Trop. Med. Hyg. 2016, 95, 938–944. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-J.; Huang, M.-C.; Wang, S.-M.; Wu, J.-J.; Cheng, C.-P.; Liu, C.-C. Analysis of Risk Factors for Bacteremia in Children with Nontyphoidal Salmonella Gastroenteritis. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; McCormick, B.A. Mucosal Inflammatory Response to Salmonella Typhimurium Infection. Front. Immunol. 2014, 5, 311. [Google Scholar] [CrossRef]
- Splichal, I.; Rychlik, I.; Splichalova, I.; Karasova, D.; Splichalova, A. Toll-Like Receptor 4 Signaling in the Ileum and Colon of Gnotobiotic Piglets Infected with Salmonella Typhimurium or Its Isogenic ∆rfa Mutants. Toxins 2020, 12, 545. [Google Scholar] [CrossRef]
- Jones, T.F.; Ingram, L.A.; Cieslak, P.R.; Vugia, D.J.; Tobin-D’Angelo, M.; Hurd, S.; Medus, C.; Cronquist, A.; Angulo, F.J. Salmonellosis Outcomes Differ Substantially by Serotype. J. Infect. Dis. 2008, 198, 109–114. [Google Scholar] [CrossRef]
- Medalla, F.; Gu, W.; Friedman, C.R.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Increased Incidence of Antimicrobial-Resistant Nontyphoidal Salmonella Infections, United States, 2004–2016. Emerg. Infect. Dis. 2021, 27, 1662–1672. [Google Scholar] [CrossRef]
- Buzilă, E.R.; Gatej, R.; Trifan, C.; Vremera, T.; Leustean, M.; David, A.; Bosogea, D.C.; Barbu, G.; Gatea, A.; Ilie, C.; et al. Genetic Characterization of Salmonella and Analysis of Ciprofloxacin Resistance Using Sanger Technique in Romania, 2024. Bacteria 2025, 4, 43. [Google Scholar] [CrossRef]
- Cao, G.; Zhao, S.; Kuang, D.; Hsu, C.-H.; Yin, L.; Luo, Y.; Chen, Z.; Xu, X.; Strain, E.; McDermott, P.; et al. Geography Shapes the Genomics and Antimicrobial Resistance of Salmonella Enterica Serovar Enteritidis Isolated from Humans. Sci. Rep. 2023, 13, 1331. [Google Scholar] [CrossRef]
- Marino, A.; Maniaci, A.; Lentini, M.; Ronsivalle, S.; Nunnari, G.; Cocuzza, S.; Parisi, F.M.; Cacopardo, B.; Lavalle, S.; La Via, L. The Global Burden of Multidrug-Resistant Bacteria. Epidemiologia 2025, 6, 21. [Google Scholar] [CrossRef]
- Marino, A.; Augello, E.; Bellanca, C.M.; Cosentino, F.; Stracquadanio, S.; La Via, L.; Maniaci, A.; Spampinato, S.; Fadda, P.; Cantarella, G.; et al. Antibiotic Therapy Duration for Multidrug-Resistant Gram-Negative Bacterial Infections: An Evidence-Based Review. Int. J. Mol. Sci. 2025, 26, 6905. [Google Scholar] [CrossRef]
- Mihaiu, L.; Lapusan, A.; Tanasuica, R.; Sobolu, R.; Mihaiu, R.; Oniga, O.; Mihaiu, M. First Study of Salmonella in Meat in Romania. J. Infect. Dev. Ctries. 2014, 8, 50–58. [Google Scholar] [CrossRef]
- Balea, L.B.; Glasdam, S. Practices, Strategies, and Challenges in Antibiotic Treatment and Prevention of Antimicrobial Resistance from the Perspectives of Romanian Community Pharmacists and General Practitioners: A Goffman-Inspired Qualitative Interview Study. Front. Antibiot. 2024, 3, 1439688. [Google Scholar] [CrossRef]
- Turgeon, P.; Murray, R.; Nesbitt, A. Hospitalizations Associated with Salmonellosis among Seniors in Canada, 2000–2010. Epidemiol. Infect. 2017, 145, 1527–1534. [Google Scholar] [CrossRef]
- Lin, R.J.; Evans, A.T.; Chused, A.E.; Unterbrink, M.E. Anemia in General Medical Inpatients Prolongs Length of Stay and Increases 30-Day Unplanned Readmission Rate. South. Med. J. 2013, 106, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Katiyo, S.; Muller-Pebody, B.; Minaji, M.; Powell, D.; Johnson, A.P.; De Pinna, E.; Day, M.; Harris, R.; Godbole, G. Epidemiology and Outcomes of Nontyphoidal Salmonella Bacteremias from England, 2004 to 2015. J. Clin. Microbiol. 2019, 57, e01189-18. [Google Scholar] [CrossRef]
- Cummings, P.L.; Kuo, T.; Javanbakht, M.; Shafir, S.; Wang, M.; Sorvillo, F. Salmonellosis Hospitalizations in the United States: Associated Chronic Conditions, Costs, and Hospital Outcomes, 2011, Trends 2000–2011. Foodborne Pathog. Dis. 2016, 13, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Pagani, G.; Parenti, M.; Franzetti, M.; Pezzati, L.; Bassani, F.; Osnaghi, B.; Vismara, L.; Pavia, C.; Mirri, P.; Rusconi, S. Invasive and Non-Invasive Human Salmonellosis Cases Admitted between 2015 and 2021 in Four Suburban Hospitals in the Metropolitan Area of Milan (Italy): A Multi-Center Retrospective Study. Pathogens 2023, 12, 1298. [Google Scholar] [CrossRef]
- Mughini-Gras, L.; Pijnacker, R.; Duijster, J.; Heck, M.; Wit, B.; Veldman, K.; Franz, E. Changing Epidemiology of Invasive Non-Typhoid Salmonella Infection: A Nationwide Population-Based Registry Study. Clin. Microbiol. Infect. 2020, 26, e9–e941. [Google Scholar] [CrossRef]
- Sia, S.; Ablola, F.; Lagrada, M.; Olorosa, A.; Gayeta, J.; Limas, M.; Jamoralin, M., Jr.; Macaranas, P.K.; Espiritu, H.G.; Borlaza, J.J.; et al. Epidemiology and Antimicrobial Resistance Profile of Invasive Non-Typhoidal Salmonella from the Philippines Antimicrobial Resistance Surveillance Program, 2014–2018. West. Pac. Surveill. Response J. 2023, 14, 23–29. [Google Scholar] [CrossRef]
- Lunguya, O.; Lejon, V.; Phoba, M.-F.; Bertrand, S.; Vanhoof, R.; Glupczynski, Y.; Verhaegen, J.; Muyembe-Tamfum, J.-J.; Jacobs, J. Antimicrobial Resistance in Invasive Non-Typhoid Salmonella from the Democratic Republic of the Congo: Emergence of Decreased Fluoroquinolone Susceptibility and Extended-Spectrum Beta Lactamases. PLoS Negl. Trop. Dis. 2013, 7, e2103. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Shan, Q.; Qiu, Y.; Lin, X.; Zhu, C.; Zhuo, Z.; Wang, C.; Tong, J.; Li, R.; Wan, C.; et al. Clinical Profiles and Antimicrobial Resistance Patterns of Invasive Salmonella Infections in Children in China. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 1215–1225. [Google Scholar] [CrossRef]
- Israel, Y.; Muhsen, K.; Rokney, A.; Adler, A. Epidemiological and Clinical Characteristics of Non-Typhoidal Salmonella Bloodstream Infections in Central Israel: A Case-Control Study. Microorganisms 2022, 10, 1942. [Google Scholar] [CrossRef]
- Björklund, L.; Mattisson, Y.; Bläckberg, A.; Sunnerhagen, T.; Ljungquist, O. A Population-Based Study on the Incidence, Risk Factors, and Outcome of Salmonella Bloodstream Infections in South Sweden 2012–2022. Infect. Dis. Ther. 2024, 13, 501–519. [Google Scholar] [CrossRef]
- Haselbeck, A.H.; Panzner, U.; Im, J.; Baker, S.; Meyer, C.G.; Marks, F. Current Perspectives on Invasive Nontyphoidal Salmonella Disease. Curr. Opin. Infect. Dis. 2017, 30, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-W.; Chen, P.-L.; Lee, N.-Y.; Lee, H.-C.; Chang, C.-M.; Lee, C.-C.; Ko, W.-C. Non-Typhoidal Salmonella Bacteremia among Adults: An Adverse Prognosis in Patients with Malignancy. J. Microbiol. Immunol. Infect. 2012, 45, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Szvalb, A.D.; Adachi, J.A.; Tarrand, J.J.; Mulanovich, V.E. Clinical Presentation and Outcomes of Non-Typhoidal Salmonella Infections in Patients with Cancer. BMC Infect. Dis. 2021, 21, 1021. [Google Scholar] [CrossRef]
- Hsu, R.-B.; Chen, R.J.; Lin, F.-Y.; Chu, S.-H. Influence of Ciprofloxacin Resistance on Risk Factors for Endovascular Infection in Patients with Infection Due to Group C Nontyphoid Salmonellae. Clin. Infect. Dis. 2005, 40, 1364–1367. [Google Scholar] [CrossRef] [PubMed]
- Wetchasirigul, S.; Puangseree, J.; Angkititrakul, S.; Prathan, R.; Srisanga, S.; Chuanchuen, R. Antimicrobial Resistance and Plasmid-Associated Virulence Genes in Salmonella Isolated from Pigs, Pork, and Humans in Border Provinces of Thailand and Neighboring Countries. PeerJ 2025, 13, e19884. [Google Scholar] [CrossRef] [PubMed]
- Khajanchi, B.K.; Foley, S.L. Antimicrobial Resistance and Increased Virulence of Salmonella. Microorganisms 2022, 10, 1829. [Google Scholar] [CrossRef] [PubMed]


| Parameter | 2018–2024 |
|---|---|
| Number of patients (n) | 698 |
| Sex (n, %) | |
| Male | 367 (52.6%) |
| Female | 331 (47.4%) |
| Residence (n, %) | |
| Urban | 289 (41.4%) |
| Rural | 409 (58.6%) |
| Age (years) | |
| Median (IQR: Q1–Q3) | 25 (IQR: 10–50.25) |
| <18 years (n, %) | 252 (36.1%) |
| ≥18 years (n, %) | 446 (63.9%) |
| Length of hospital stay (days) | |
| Median (IQR: Q1–Q3) | 5 (IQR: 3–6) |
| Comorbidities (n, %) | |
| Diabetes mellitus | 40 (5.7%) |
| Cardiovascular disease | 117 (16.8%) |
| Pulmonary disease | 21 (3%) |
| Malignancy (all types) | 15 (2.1%) |
| Parameter | All Infections | Serogroup B | Serogroup C | Serogroup D | Normal Range |
|---|---|---|---|---|---|
| WBC * (cells/mm3) | 4000–10,000 | ||||
| Median (IQR: Q1–Q3) | 7750 (IQR: 5855–10,015) | 8175 (IQR: 6302.5–11,075) | 7969 (IQR: 5810–9715) | 7335 (IQR: 5720–9795) | |
| Neutrophils (cells/mm3) | 2000–8000 | ||||
| Median (IQR: Q1–Q3) | 5010 (IQR: 3480–7140) | 5180 (IQR: 3487.5–7150) | 5520 (IQR: 3525–6815) | 4970 (IQR: 3497.5–7242.5) | |
| RBC * (cells/mm3) | 4,400,000–5,800,000 | ||||
| Median (IQR: Q1–Q3) | 4,590,000 (IQR: 4,270,000–4,995,000) | 4,490,000 (IQR: 4,205,000–4,900,000) | 4,620,000 (IQR: 4,300,000–4,860,000) | 4,630,000 (IQR: 4,280,000–5,067,500) | |
| Hemoglobin (g/dL) | 11.7–15.5 | ||||
| Mean ± SD | 13.28 ± 1.86 | 12.96 ± 1.91 | 13.2 ± 2.06 | 13.41 ± 1.77 | |
| CRP (mg/L) | 0–5 | ||||
| Median (IQR: Q1–Q3) | 84.72 (IQR: 26.29–140.39) | 77.15 (IQR: 28.54–135.85) | 74.24 (IQR: 9.17–149.86) | 92.09 (IQR: 29.04–141.78) | |
| Serum sodium (mmol/L) | 135–148 | ||||
| Median (IQR: Q1–Q3) | 141.6 (IQR: 139.2–144) | 142.1 (IQR: 139.6–144.47) | 142.9 (IQR: 140.2–144.9) | 141.3 (IQR: 138.8–143.6) | |
| Serum potassium (mmol/L) | 3.7–5.3 | ||||
| Mean ± SD | 4.01 ± 0.50 | 4.01 ± 0.60 | 4.02 ± 0.52 | 4.01 ± 0.46 | |
| Serum chloride (mmol/L) | 98–109 | ||||
| Median (IQR: Q1–Q3) | 101.14 ± 3.24 | 101.3 (IQR: 99.3–103.52) | 101.8 (IQR: 100.7–104.4) | 101 (IQR: 99–103) |
| Serogroup | 2018 | 2019 | 2020 | 2021 | 2022 | 2023 | 2024 | Total |
|---|---|---|---|---|---|---|---|---|
| B (n, %) | 45 (17%) | 57 (36.8%) | 5 (45.5%) | 9 (32.1%) | 8 (16%) | 13 (18.8%) | 15 (12.4%) | 152 (21.8%) |
| C (n, %) | 29 (11%) | 29 (18.7%) | 2 (18.2%) | 5 (17.9%) | 13 (26%) | 9 (13%) | 11 (9.1%) | 98 (14%) |
| D (n, %) | 188 (71.2%) | 67 (43.2%) | 4 (36.4%) | 14 (50%) | 29 (58%) | 47 (68.1%) | 95 (78.5%) | 444 (63.6%) |
| E (n, %) | 2 (0.8%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (0.3%) |
| G (n, %) | 0 (0%) | 1 (0.6%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0.1) |
| UN (n, %) * | 0(0%) | 1 (0.6%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0.1%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sima, C.M.; Bădescu, A.C.; Buruiană, G.; Duhaniuc, A.; Iancu, L.S.; Năstase, E.-V.; Dorneanu, O.S. Seven Years of Salmonella: Changing Resistance and Clinical Insights. Microorganisms 2025, 13, 2655. https://doi.org/10.3390/microorganisms13122655
Sima CM, Bădescu AC, Buruiană G, Duhaniuc A, Iancu LS, Năstase E-V, Dorneanu OS. Seven Years of Salmonella: Changing Resistance and Clinical Insights. Microorganisms. 2025; 13(12):2655. https://doi.org/10.3390/microorganisms13122655
Chicago/Turabian StyleSima, Cristina Mihaela, Aida Corina Bădescu, Georgiana Buruiană, Alexandru Duhaniuc, Luminița Smaranda Iancu, Eduard-Vasile Năstase, and Olivia Simona Dorneanu. 2025. "Seven Years of Salmonella: Changing Resistance and Clinical Insights" Microorganisms 13, no. 12: 2655. https://doi.org/10.3390/microorganisms13122655
APA StyleSima, C. M., Bădescu, A. C., Buruiană, G., Duhaniuc, A., Iancu, L. S., Năstase, E.-V., & Dorneanu, O. S. (2025). Seven Years of Salmonella: Changing Resistance and Clinical Insights. Microorganisms, 13(12), 2655. https://doi.org/10.3390/microorganisms13122655





