Construction of Recombinant Escherichia coli Expressing Ammonia Assimilation Genes and Evaluation of Its Effect on Removing Ammonium Nitrogen (NH4+-N)
Abstract
1. Introduction
2. Literature Review on Biological Treatment of High-Strength Ammonia Wastewater
- (1)
- Iron-modified bentonite (f-MB) can efficiently and simultaneously remove phosphate and ammonium from various wastewaters, and nutrients can be recovered through regeneration, yielding slow-release fertilizers and enabling a shift from “pollution control” to “resource recovery” [36].
- (2)
- PhoslockTM used as a sediment capping material can sustainably inhibit endogenous phosphorus release even under conditions closer to real environments (anaerobic and in the presence of dissolved organic matter, DOM), underscoring the stability and practicality of the “chemical fixation + ecological restoration” coupling strategy for in-lake endogenous pollution control [37].
3. Materials and Methods
3.1. Bacterial Strains and Plasmid Constructions
3.2. Culture Conditions
3.3. Construction of the Recombinant Strains
3.4. In Vitro Fermentation Test Analysis
3.5. RNA Extraction, cDNA Synthesis, and RT-PCR
3.6. Water Quality Analyses
3.7. Statistical Analysis
4. Results
4.1. Cloning of Ammonia Assimilation Genes
4.2. Optimization of Factors Affecting Protein Expression
4.3. Assessment of Ammonium Removal Capacity
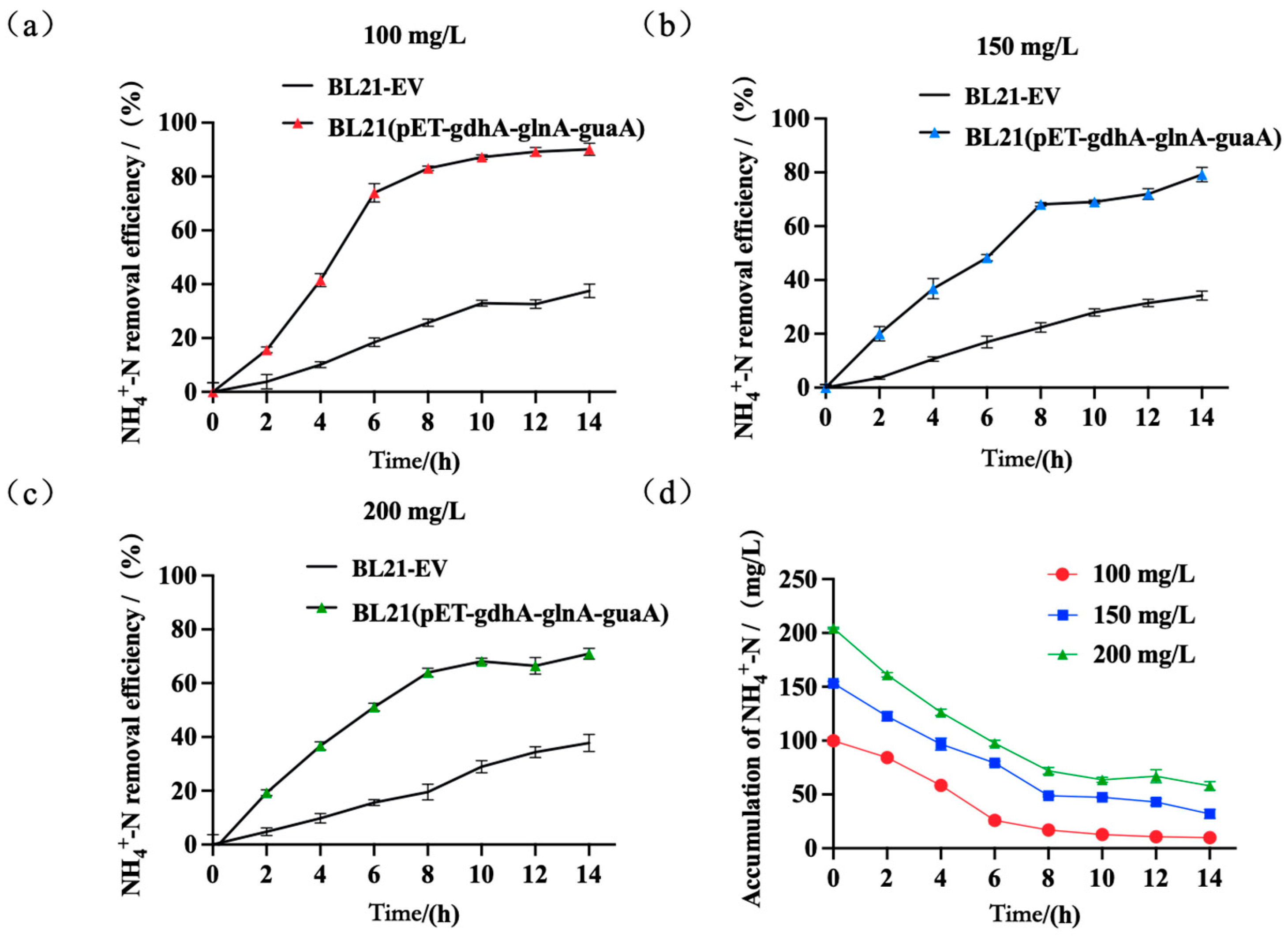
4.4. Removal of Ammonia Nitrogen at Varying Concentrations of Ammonium Nitrogen
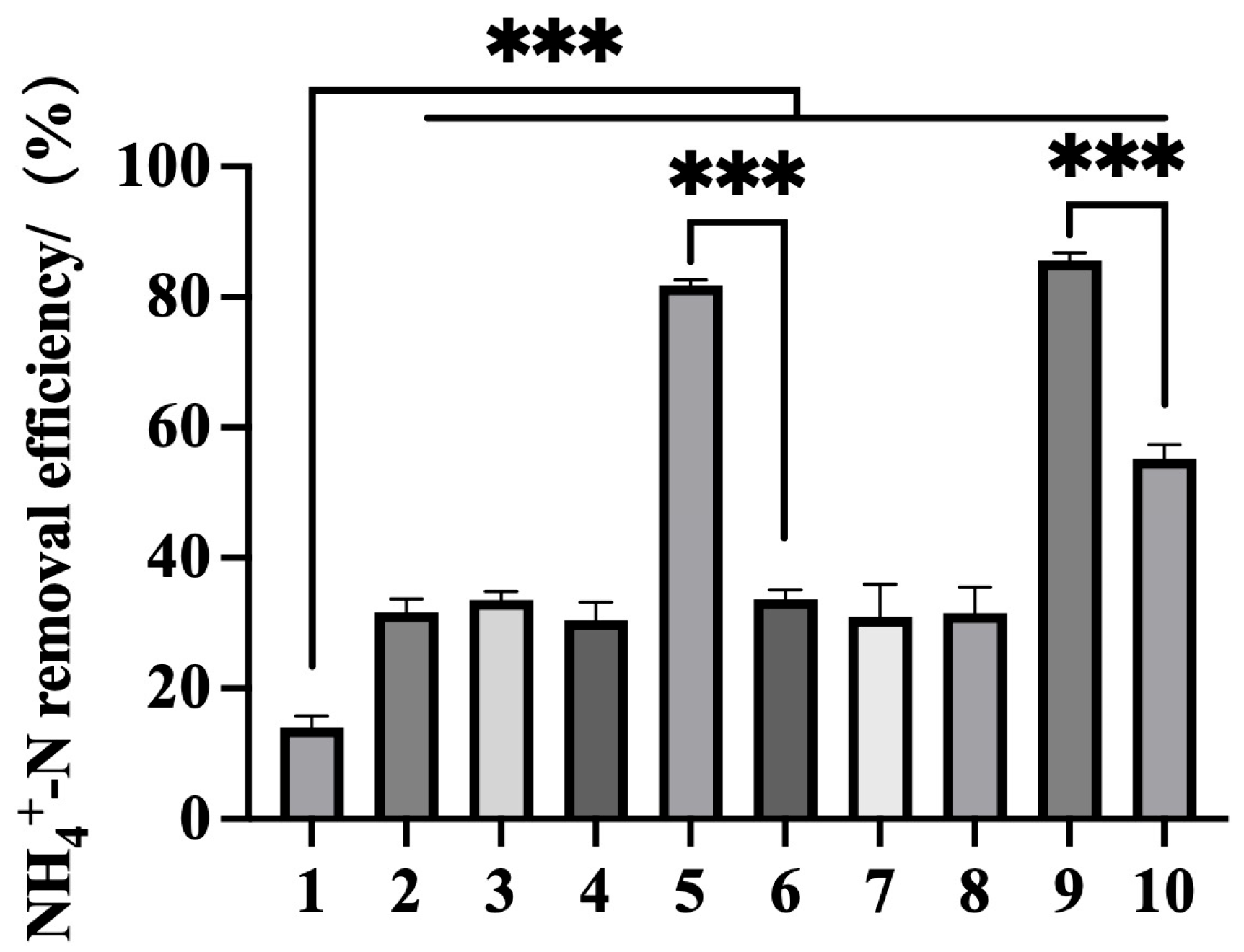
4.5. Efficient Ammonium Removal by Co-Expressed Recombinant Bacteria
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| IPTG | isopropyl β-D-1-thiogalactopyranoside |
| HNAD | heterotrophic nitrification–aerobic denitrification |
| DNRA | dissimilatory nitrate reduction to ammonium |
| GDH | glutamate dehydrogenase |
| GS | glutamine synthetase |
| GOGAT | glutamate synthase |
| COD/N | carbon/nitrogen ratio |
| PCR | polymerase chain reaction |
| LB | Luria–Bertani |
| Amp | Ampicillin |
| Kan | Kanamycin |
| RT-PCR | reverse transcription PCR |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| HRT | hydraulic retention time |
| SRT | sludge retention time |
References
- Liu, X.; Beusen, A.H.W.; van Grinsven, H.J.M.; Wang, J.; Kim, H.; van Hoek, W.J.; Ran, X.; Mogollón, J.M.; Bouwman, A.F. Impact of groundwater nitrogen legacy on water quality. Nat. Sustain. 2024, 7, 891–900. [Google Scholar] [CrossRef]
- Mishra, S.; Singh, V.; Cheng, L.; Hussain, A.; Ormeci, B. Nitrogen removal from wastewater: A comprehensive review of biological nitrogen removal processes, critical operation parameters and bioreactor design. J. Environ. Chem. Eng. 2022, 10, 107387. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, Y.; Zhu, J.; Li, C.; Chen, G. A comprehensive review on wastewater nitrogen removal and its recovery processes. Int. J. Environ. Res. Public Health 2023, 20, 3429. [Google Scholar] [CrossRef]
- Ye, Y.; Zhang, K.; Peng, X.; Zhou, Q.; Pan, Z.; Xing, B.; Liu, X. Research progress on biological denitrification process in wastewater treatment. Water 2025, 17, 520. [Google Scholar] [CrossRef]
- Kosgey, K.; Zungu, P.V.; Bux, F.; Kumari, S. Biological nitrogen removal from low carbon wastewater. Front. Microbiol. 2022, 13, 968812. [Google Scholar] [CrossRef] [PubMed]
- Owaes, M.; Gani, K.M.; Kumari, S.; Seyam, M.; Bux, F. Implementation of partial nitrification in wastewater treatment systems by modifications in operational strategies—A review. Environ. Technol. Rev. 2024, 13, 379–397. [Google Scholar] [CrossRef]
- Shi, S.; He, X.; He, L.; Fan, X.; Shu, B.; Zhou, J. Overlooked pathways of endogenous simultaneous nitrification and denitrification in anaerobic/aerobic/anoxic sequencing batch reactors with organic supplementation. Water Res. 2023, 230, 119493. [Google Scholar] [CrossRef]
- Peng, Z.; Lei, Y.; Zhan, Y.; Yang, B.; Pan, X. Impact of COD/TN ratio on shifting autotrophic partial nitrification to heterotrophic nitrification and aerobic denitrification in high-strength ammonium wastewater. Water 2024, 16, 2532. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, X.; Zhou, X.; Chen, C. Recent advances in autotrophic biological nitrogen removal for low carbon wastewater: A review. Water 2022, 14, 1101. [Google Scholar] [CrossRef]
- Msimango, S.S.; Nasr, M.; Bux, F.; Kumari, S. Impact of chemical oxygen demand to nitrogen ratio on ANAMMOX bacterial growth in an up-flow anaerobic sludge blanket reactor. Water Sci. Technol. 2024, 90, 2978–2990. [Google Scholar] [CrossRef]
- Fu, W.; Song, G.; Wang, Y.; Wang, Q.; Duan, P.; Liu, C.; Zhang, X.; Rao, Z. Advances in research into and applications of heterotrophic nitrifying and aerobic denitrifying microorganisms. Front. Environ. Sci. 2022, 10, 887093. [Google Scholar] [CrossRef]
- Zhang, M.; He, T.; Wu, Q.; Chen, M. Efficient detoxication of hydroxylamine and nitrite through heterotrophic nitrification and aerobic denitrification by Acinetobacter johnsonii EN-J1. Front. Microbiol. 2023, 14, 1130512. [Google Scholar] [CrossRef]
- Li, Q.; He, Y.; Wang, B.; Weng, N.; Zhang, L.; Wang, K.; Tian, F.; Lyu, M.; Wang, S. Heterotrophic nitrification–aerobic denitrification by Bacillus sp. L2: Mechanism of denitrification and strain immobilization. Water 2024, 16, 416. [Google Scholar] [CrossRef]
- Ren, J.; Tang, J.; Min, H.; Tang, D.; Jiang, R.; Liu, Y.; Huang, X. Nitrogen removal characteristics of novel bacterium Klebsiella sp. TSH15 by assimilatory/dissimilatory nitrate reduction and ammonia assimilation. Bioresour. Technol. 2024, 394, 130184. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Luo, Y.; Luo, L.; Xie, D.; Li, Z. The nitrite reductase encoded by nirBDs in Pseudomonas putida Y-9 influences ammonium transformation. Front. Microbiol. 2022, 13, 982674. [Google Scholar] [CrossRef]
- Tecson, M.C.B.; Geluz, C.; Cruz, Y.; Greene, E.R. Glutamine Synthetase: Diverse Regulation and Functions of an Ancient Enzyme. Biochemistry 2025, 64, 547–554. [Google Scholar] [CrossRef]
- Harling, L.C.; Hecht, A.L.; Meyer, F.; Wu, G.D. Revisiting nitrogen assimilation strategies in the mammalian gut: Lessons from Enterobacteriaceae as pathobiont models and a challenge to the limitation paradigm. Arch. Microbiol. 2025, 207, 203. [Google Scholar] [CrossRef]
- Williamson, G.; Bizior, A.; Harris, T.; Pritchard, L.; Hoskisson, P.A.; Javelle, A. Biological ammonium transporters from the Amt/Mep/Rh superfamily: Mechanism, energetics, and technical limitations. Biosci. Rep. 2024, 44, BSR20211209. [Google Scholar] [CrossRef]
- Chi, Z.; Li, Y.; Zhang, J.; Hu, M.; Wu, Y.; Fan, X.; Li, Z.; Miao, Q.; Li, W. Effects of nitrogen application on ammonia assimilation and microenvironment in the rhizosphere of drip-irrigated sunflower under plastic mulch. Front. Microbiol. 2024, 15, 1390331. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, Y.; Guan, W.; Qi, W.; Tai, X.; Lin, D.; He, R.; Sun, L. Research on nitrogen transformation pathways of a thermophilic heterotrophic nitrifying bacterial consortium GW7. Front. Microbiol. 2025, 16, 1578865. [Google Scholar] [CrossRef]
- Sun, W.; Hu, C.; Wu, J.; Wei, M.; Lin, J.G.; Hong, Y. Efficient nitrogen removal via simultaneous ammonia assimilation and heterotrophic denitrification of Paracoccus denitrificans R-1. iScience 2024, 27, 110599. [Google Scholar] [CrossRef]
- Ge, F.; Sun, J.; Ren, Y.; He, B.; Li, J.; Yang, S.; Li, W. Transcriptomic and enzymatic analysis reveals the roles of glutamate dehydrogenases GdhA/GdhB in nitrogen–carbon coupling. AMB Express 2022, 12, 161. [Google Scholar] [CrossRef]
- Wang, F.; Lv, X.; Guo, Z.; Wang, X.; Long, Y.; Liu, H. Functional Characterization of Two Glutamate Dehydrogenase Genes in Bacillus altitudinis AS19 and Optimization of Soluble Recombinant Expression. Curr. Issues Mol. Biol. 2025, 47, 603. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zheng, N.; Wang, J.; Zhao, S. Nitrogen metabolism of the highly ureolytic bacterium Proteus penneri S99 isolated from the rumen. BMC Microbiol. 2025, 25, 104. [Google Scholar] [CrossRef] [PubMed]
- Valderrama-Martín, J.M.; Ortigosa, F.; Ávila, C. A revised view on the evolution of glutamine synthetase isoenzymes in plants. Plant J. 2022, 110, 946–960. [Google Scholar] [CrossRef]
- Travis, B.A.; Peck, J.V.; Salinas, R.; Dopkins, B.; Lent, N.; Nguyen, V.D.; Borgnia, M.J.; Brennan, R.G.; Schumacher, M.A. Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria. Nat. Commun. 2022, 13, 3793. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.C.; Lemaire, O.N.; Kurth, J.M.; Welte, C.U.; Wagner, T. Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations. Commun. Biol. 2024, 7, 111. [Google Scholar] [CrossRef]
- Herdering, E.; Reif-Trauttmansdorff, T.; Kumar, A.; Habenicht, T.; Hochberg, G.; Bohn, S.; Schuller, J.; Schmitz, R.A.; Ropers, D. 2-Oxoglutarate triggers assembly of active dodecameric glutamine synthetase. eLife 2025, 14, e97484. [Google Scholar] [CrossRef]
- Huang, L.; Lu, W.; Yu, Y.; Qiu, H.; Zeng, Y.; Wang, L.; Liu, Y.; Yan, L.; Fu, Y.V.; Zheng, Y. The ammonium transporter AmtB is dispensable for the uptake of ammonium in the phototrophic diazotroph Rhodopseudomonas palustris. Environ. Technol. Innov. 2024, 36, 103853. [Google Scholar] [CrossRef]
- Williamson, G.; Brito, A.S.; Bizior, A.; Tamburrino, G.; Dias Mirandela, G.; Harris, T.; Hoskisson, P.A.; Zachariae, U.; Marini, A.M.; Boeckstaens, M.; et al. Coexistence of ammonium transporter and channel ensures efficient NH4+ scavenging. mBio 2022, 13, e0291321. [Google Scholar] [CrossRef]
- Han, F.; Zhang, M.; Li, Z.; Liu, Z.; Han, Y.; Li, Y.; Zhou, W. Self-assembly of ammonia assimilation microbiomes regulated by COD/N ratio. Chem. Eng. J. 2023, 455, 140782. [Google Scholar] [CrossRef]
- Guo, Y.; Han, F.; Zhang, M.; Zhao, C.; Zhou, W. Fast nutrients removal through strengthened solid-liquid conversion and biomass synthesis of heterotrophic ammonia assimilation system in saline wastewater. Environ. Res. 2025, 282, 122050. [Google Scholar] [CrossRef]
- Liu, R.; Qin, H.; Wang, Q.; Chu, C.; Jiang, Y.; Deng, H.; Han, C.; Zhong, W. Transcriptome Analysis of Nitrogen Assimilation Preferences in Burkholderia sp. M6-3 and Arthrobacter sp. M7-15. Front. Microbiol. 2025, 16, 1559884. [Google Scholar] [CrossRef] [PubMed]
- Zhen, J.; Zhao, Y.; Yu, X.; Guo, W.; Qiao, Z.; Ismail, S.; Ni, S.-Q. Feasibility of Partial Nitrification Combined with Nitrite-Denitrification Phosphorus Removal and Simultaneous Nitrification–Endogenous Denitrification for Synchronous Chemical Oxygen Demand, Nitrogen, and Phosphorus Removal. ACS EST Water 2022, 2, 1119–1131. [Google Scholar] [CrossRef]
- Chen, J.; Xu, J.; Zhang, S.; Liu, F.; Peng, J.; Peng, Y.; Wu, J. Nitrogen Removal Characteristics of a Novel Heterotrophic Nitrification and Aerobic Denitrification Bacteria, Alcaligenes Faecalis Strain WT14. J. Environ. Manag. 2021, 282, 111961. [Google Scholar] [CrossRef]
- Zamparas, M.; Kyriakopoulos, G.L.; Drosos, M.; Kapsalis, V.C. Phosphate and Ammonium Removal from Wastewaters Using Natural-Based Innovative Bentonites Impacting on Resource Recovery and Circular Economy. Molecules 2021, 26, 6684. [Google Scholar] [CrossRef]
- Zamparas, M.; Kyriakopoulos, G.L.; Kapsalis, V.C.; Drosos, M.; Kalavrouziotis, I.K. Application of Novel Composite Materials as Sediment Capping Agents: Column Experiments and Modelling. Desalination Water Treat. 2019, 170, 111–118. [Google Scholar] [CrossRef]
- Xiao, R.; Tian, C.; Wang, H.; Zhang, H.; Chen, H.; Chou, H.H. Two-stage continuous cultivation of microalgae overexpressing cytochrome P450 improves nitrogen and antibiotics removal from livestock and poultry wastewater. Bioresour. Technol. 2025, 418, 131994. [Google Scholar] [CrossRef]
- Pahlevan Kakhki, M. TRIzol-based RNA extraction: A reliable method for gene expression studies. J. Sci. Islam. Repub. Iran 2014, 25, 13–17. [Google Scholar]
- Ivarsson, K.; Weijdegård, B. Evaluation of the effects of DNase treatment on signal specificity in RT-PCR and in situ RT-PCR. Biotechniques 1998, 25, 630–638. [Google Scholar] [CrossRef]
- Bachman, J. Reverse-transcription PCR (rt-PCR). Methods Enzymol. 2013, 530, 67–74. [Google Scholar]
- HJ 535-2009; Water Quality—Determination of Ammonia Nitrogen—Nessler’s Reagent Spectrophotometry. China Environmental Science Press: Beijing, China, 2009.
- Zhang, H.; Yang, Z.; Tian, J.; Liu, C.; Qin, Z. Enhanced Nitrate Nitrogen Removal from Wastewater Using Modified Reed Straw: Adsorption Performance and Resource Utilization. Sustainability 2024, 16, 4001. [Google Scholar] [CrossRef]
- Williams, L.J.; Abdi, H. Post-hoc comparisons. Encycl. Res. Des. 2010, 1060–1067. Available online: https://personal.utdallas.edu/~Herve/abdi-PostHoc2010-pretty.pdf (accessed on 9 October 2025).
- Elliott, A.C.; Hynan, L.S. A SAS® macro implementation of a multiple comparison post hoc test for a Kruskal–Wallis analysis. Comput. Methods Programs Biomed. 2011, 102, 75–80. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, H.; Zhu, R.; Liao, X.; Wu, Y.; Mi, J.; Wang, Y. Ammonia reduction by the gdhA and glnA genes from bacteria in laying hens. Ecotoxicol. Environ. Saf. 2021, 222, 112486. [Google Scholar] [CrossRef] [PubMed]
- Urs, K.; Zimmern, P.E.; Reitzer, L. Control of glnA (glutamine synthetase) expression by urea in non-pathogenic and uropathogenic Escherichia coli. J. Bacteriol. 2023, 205, e0026823. [Google Scholar] [CrossRef]
- Mühlmann, M.; Forsten, E.; Noack, S.; Büchs, J. Optimizing recombinant protein expression via automated induction profiling in microtiter plates at different temperatures. Microb. Cell Factories 2017, 16, 131. [Google Scholar] [CrossRef]
- Horga, L.G.; Halliwell, S.; Castiñeiras, T.S.; Wyre, C.; Matos, C.F.R.O.; Yovchaeva, D.S.; Kent, R. Tuning recombinant protein expression to match secretion capacity. Microb. Cell Factories 2018, 17, 199. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Wang, W.; Rong, J.; Liang, J.; Mi, J.; Wu, Y.; Wang, Y. Construction of recombinant Pichia pastoris strains for ammonia reduction by the gdhA and glnA regulatory genes in laying hens. Ecotoxicol. Environ. Saf. 2022, 234, 113376. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.L.; Duan, P.F.; Wang, Q.; Liao, Y.; Wang, Y.; Xu, M.; Jiang, H.; Zhang, X.; Rao, Z. Transcriptomics Reveals the Effect of Ammonia Nitrogen Concentration on Pseudomonas Stutzeri F2 Assimilation and the Analysis of amtB Function. Synth. Syst. Biotechnol. 2023, 8, 262–272. [Google Scholar] [CrossRef]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martínez, R.A.; Martínez Vivancos, A.; Cánovas Díaz, M.; de Diego Puente, T. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef]
- Zhao, C.; Li, X.; Guo, L. Reprogramming Metabolic Flux in Escherichia coli to Enhance Chondroitin Production. Adv. Sci. 2023, 11, e2307351. [Google Scholar] [CrossRef]
- Nan, J.; Zhang, S.; Zhan, P.; Jiang, L. Discovery of Novel GMPS Inhibitors of Candidatus Liberibacter Asiaticus by Structure Based Design and Enzyme Kinetic. Biology 2021, 10, 594. [Google Scholar] [CrossRef]
- Barrick, J.E.; Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007, 8, R239. [Google Scholar] [CrossRef]
- Qin, Y.L.; Liang, Z.L.; Ai, G.M.; Liu, W.F.; Tao, Y.; Jiang, C.Y.; Liu, S.J.; Li, D.F. Heterotrophic nitrification by Alcaligenes faecalis links organic and inorganic nitrogen metabolism. ISME J. 2024, 18, wrae174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, K.; Liu, X.; Chen, H.; Cai, Z. Treatment of High-Ammonia-Nitrogen Wastewater with Immobilized Ammonia-Oxidizing Bacteria Alcaligenes sp. TD-94 and Paracoccus sp. TD-10. Processes 2023, 11, 926. [Google Scholar] [CrossRef]
- Liu, Y.; Ngo, H.H.; Guo, W.; Peng, L.; Wang, D.; Ni, B. The roles of free ammonia (FA) in biological wastewater treatment processes: A review. Environ. Int. 2019, 123, 10–19. [Google Scholar] [CrossRef] [PubMed]
- van Heeswijk, W.C.; Westerhoff, H.V.; Boogerd, F.C. Nitrogen assimilation in Escherichia coli: Putting molecular data into a physiological context. Microbiol. Mol. Biol. Rev. 2013, 77, 628–695. [Google Scholar] [CrossRef]
- Kumar, R.; Shimizu, K. Metabolic regulation of Escherichia coli and its gdhA, glnL, gltB, D mutants under different carbon and nitrogen limitations in the continuous culture. Microb. Cell Factories 2010, 9, 8. [Google Scholar] [CrossRef]
- Rahimi, S.; Modin, O.; Mijakovic, I. Technologies for biological removal and recovery of nitrogen from wastewater. Biotechnol. Adv. 2020, 43, 107570. [Google Scholar] [CrossRef]
- Thurston, R.V. Aqueous Ammonia Equilibrium: Tabulation of Percent Un-Ionized Ammonia; Environmental Research Laboratory-Duluth, Office of Research and Development, US Environmental Protection Agency, University of California Riverside: Washington, DC, USA, 1979. [Google Scholar]
- Martín-Rodríguez, A.J.; Rhen, M.; Melican, K.; Richter-Dahlfors, A. Nitrate Metabolism Modulates Biosynthesis of Biofilm Components in Uropathogenic Escherichia coli and Acts as a Fitness Factor During Experimental Urinary Tract Infection. Front. Microbiol. 2020, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Waqas, S.; Harun, N.Y.; Sambudi, N.S. Effect of Operating Parameters on the Performance of Integrated Fixed-Film Activated Sludge for Wastewater Treatment. Membranes 2023, 13, 704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, W.; Li, D.; Wang, S.; Lv, L. Integration of ammonium assimilation with denitrifying phosphorus removal for efficient nutrient management in wastewater treatment. J. Environ. Manag. 2024, 353, 120116. [Google Scholar] [CrossRef]
- Ma, H.; Zhao, Y.; Yang, K.; Wang, Y.; Zhang, C.; Ji, M. Application oriented bioaugmentation processes: Mechanism, performance improvement and scale-up. Bioresour. Technol. 2022, 344, 126192. [Google Scholar] [CrossRef]
- Zhao, Z.; Cheng, J.F.; Yoshikuni, Y. Chromosomal integration of complex DNA constructs using CRAGE and CRAGE-Duet systems. STAR Protoc. 2022, 3, 101546. [Google Scholar] [CrossRef]
- Brechun, K.E.; Förschle, M.; Schmidt, M.; Kranz, H. Method for plasmid-based antibiotic-free fermentation. Microb. Cell Factories 2024, 23, 18. [Google Scholar] [CrossRef]
- Izadi, P.; Izadi, P.; Eldyasti, A. Towards mainstream deammonification: Comprehensive review on potential mainstream applications and developed sidestream technologies. J. Environ. Manag. 2021, 279, 111615. [Google Scholar] [CrossRef] [PubMed]
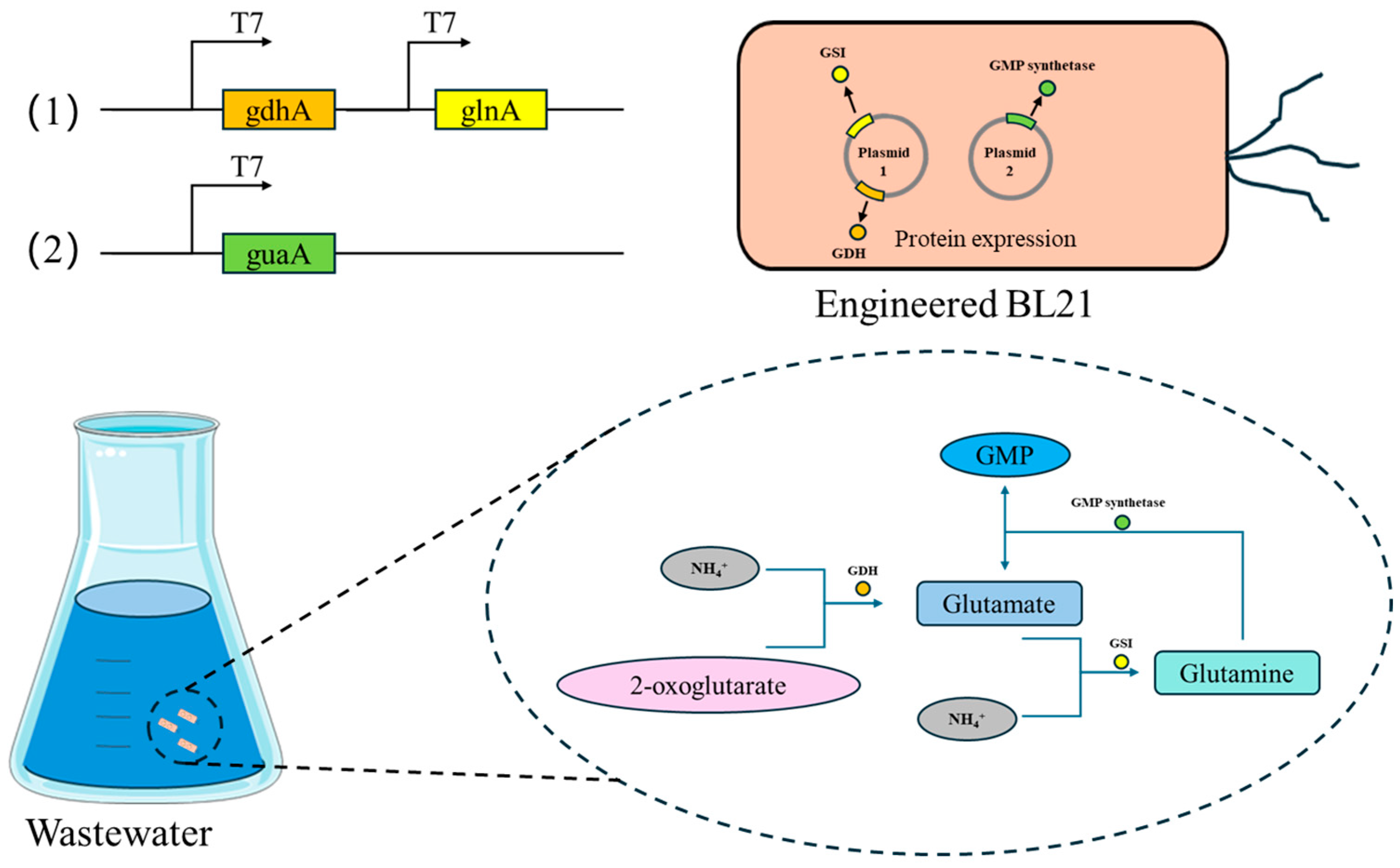

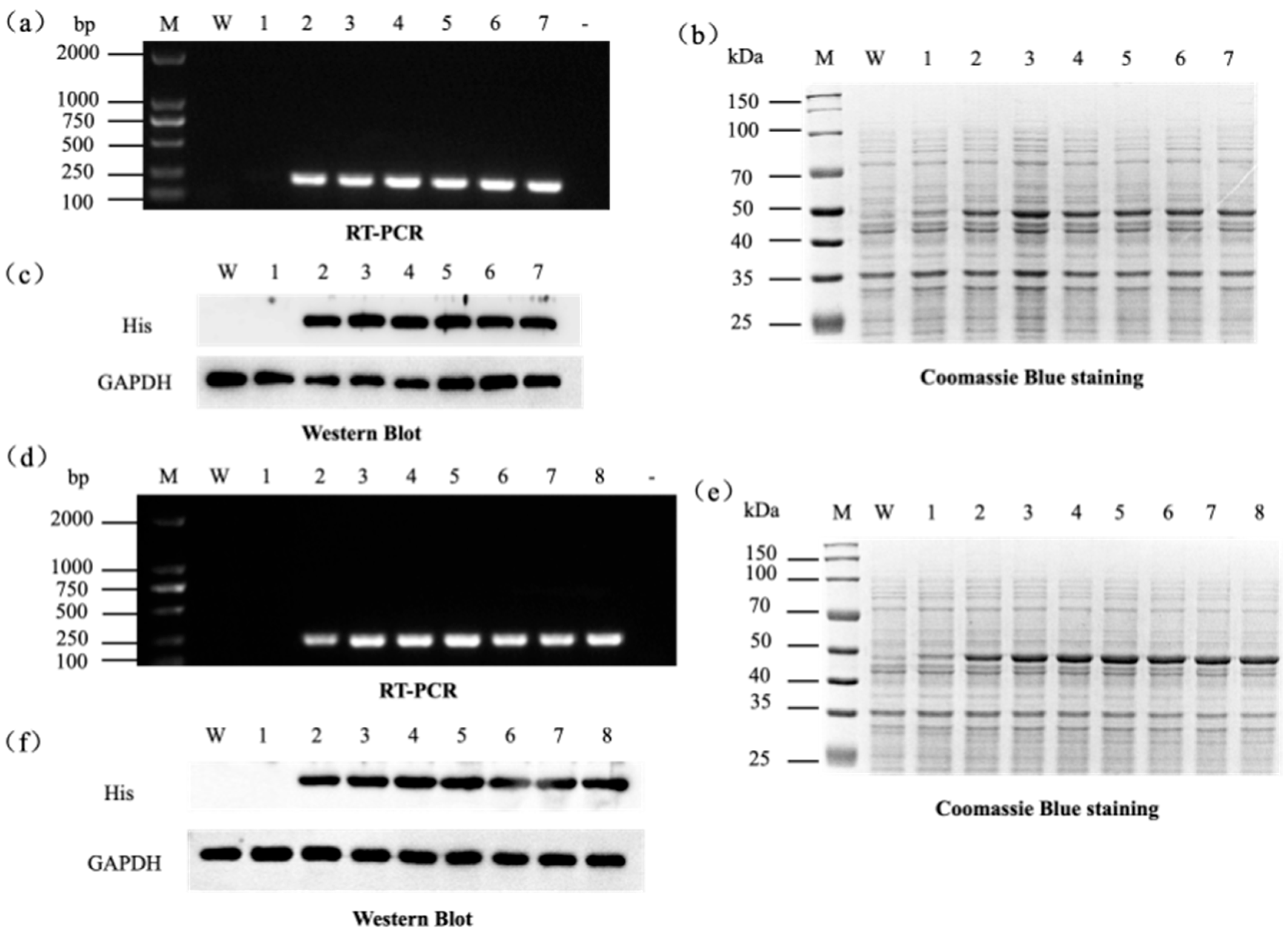

| Gene | Organism (Strain) | Sequence Accession (RefSeq/GenBank) | Protein Length (aa) |
|---|---|---|---|
| gdhA | Enterococcus faecium | WP_002317577.1 (RefSeq protein); genomic NZ_CP038996.1: complement(1,669,435..1,670,784); NCBI Gene ID: 66454627 | 449 |
| glnA | Heyndrickxia coagulans DSM 1 | WP_029141484.1 (RefSeq protein); genome NZ_CP009709.1, complement(547,556..548,893); NCBI Gene ID: 29813122 | 445 |
| guaA | Heyndrickxia coagulans DSM 1 | WP_029142766.1 (RefSeq protein); genome NZ_CP009709.1, complement(1,890,032..1,891,585); NCBI Gene ID: 29811950 | 517 |
| Strain Name | Expression Vector | Expressed Genes | Gene Functions |
|---|---|---|---|
| BL21(pET-gdhA) | pET-28a(+) | gdhA | Encodes glutamate dehydrogenase, involved in ammonia assimilation. |
| BL21(pET-glnA) | pET-28a(+) | glnA | Encodes glutamine synthetase (GS), catalyzes the synthesis of glutamine from glutamate and ammonia. |
| BL21(pET-guaA) | pET-28a(+) | guaA | Encodes GMP synthetase (GMPS), involved in guanosine synthesis. |
| BL21(pET-gdhA-glnA) | pETDuet-1 | gdhA, glnA | Encodes both glutamate dehydrogenase and glutamine synthetase. |
| BL21(pET-glnA-guaA) | pETDuet-1 | glnA, guaA | Encodes both glutamine synthetase and GMP synthetase. |
| BL21(pET-gdhA-glnA-guaA) * | pETDuet1+ pET-28a(+) | gdhA, glnA, guaA | Encodes glutamate dehydrogenase, glutamine synthetase and GMP synthetase. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, P.; Yang, Y.; Shi, R.; Kang, Y.; Xu, H.; Cheng, X.; Yan, Q.; Hu, H. Construction of Recombinant Escherichia coli Expressing Ammonia Assimilation Genes and Evaluation of Its Effect on Removing Ammonium Nitrogen (NH4+-N). Microorganisms 2025, 13, 2646. https://doi.org/10.3390/microorganisms13122646
Pan P, Yang Y, Shi R, Kang Y, Xu H, Cheng X, Yan Q, Hu H. Construction of Recombinant Escherichia coli Expressing Ammonia Assimilation Genes and Evaluation of Its Effect on Removing Ammonium Nitrogen (NH4+-N). Microorganisms. 2025; 13(12):2646. https://doi.org/10.3390/microorganisms13122646
Chicago/Turabian StylePan, Pan, Yongkun Yang, Runxuan Shi, Yulin Kang, Hanli Xu, Xiyu Cheng, Qiong Yan, and Honggang Hu. 2025. "Construction of Recombinant Escherichia coli Expressing Ammonia Assimilation Genes and Evaluation of Its Effect on Removing Ammonium Nitrogen (NH4+-N)" Microorganisms 13, no. 12: 2646. https://doi.org/10.3390/microorganisms13122646
APA StylePan, P., Yang, Y., Shi, R., Kang, Y., Xu, H., Cheng, X., Yan, Q., & Hu, H. (2025). Construction of Recombinant Escherichia coli Expressing Ammonia Assimilation Genes and Evaluation of Its Effect on Removing Ammonium Nitrogen (NH4+-N). Microorganisms, 13(12), 2646. https://doi.org/10.3390/microorganisms13122646






