-
 MTHFR Gene Polymorphisms: A Single Gene with Wide-Ranging Clinical Implications—A Review
MTHFR Gene Polymorphisms: A Single Gene with Wide-Ranging Clinical Implications—A Review -
 Role of MicroRNAs in Acute Myeloid Leukemia
Role of MicroRNAs in Acute Myeloid Leukemia -
 SYNGAP1 Syndrome and the Brain Gene Registry
SYNGAP1 Syndrome and the Brain Gene Registry -
 Genetics of Suicide
Genetics of Suicide -
 Genetic Features of Tumours Arising in the Context of Suspected Hereditary Cancer Syndromes with RAD50, RAD51C/D, and BRIP1 Germline Mutations, Results of NGS-Reanalysis of BRCA/MMR-Negative Families
Genetic Features of Tumours Arising in the Context of Suspected Hereditary Cancer Syndromes with RAD50, RAD51C/D, and BRIP1 Germline Mutations, Results of NGS-Reanalysis of BRCA/MMR-Negative Families
Journal Description
Genes
Genes
is a peer-reviewed, open access journal of genetics and genomics published monthly online by MDPI. The Spanish Society for Biochemistry and Molecular Biology (SEBBM) is affiliated with Genes and their members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Genetics and Heredity) / CiteScore - Q2 (Genetics (clinical))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 14.9 days after submission; acceptance to publication is undertaken in 2.6 days (median values for papers published in this journal in the second half of 2024).
- Recognition of Reviewers: Reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
2.8 (2024);
5-Year Impact Factor:
3.2 (2024)
Latest Articles
Using STR Data to Investigate the Impact of the Studbook Cap on Genetic Diversity in the American Standardbred Horse from 1998 to 2021
Genes 2025, 16(7), 748; https://doi.org/10.3390/genes16070748 (registering DOI) - 27 Jun 2025
Abstract
Background/Objectives: Standardbreds, a breed of horses used in harness racing at either the trot or the pace, established a closed studbook in 1973. Concerns about genetic diversity within the breed led the United States Trotting Association (USTA) to establish a limit of mares
[...] Read more.
Background/Objectives: Standardbreds, a breed of horses used in harness racing at either the trot or the pace, established a closed studbook in 1973. Concerns about genetic diversity within the breed led the United States Trotting Association (USTA) to establish a limit of mares bred per stallion (i.e., a studbook cap) in 2009. Here, we aimed to evaluate the impact of the breeding restrictions on genetic diversity between and among subpopulations. Methods: Sixteen short tandem repeats (STRs) were analyzed across a dataset of 176,424 Standardbreds foaled in the United States between 1998 and 2021. We examined allelic richness (Na), number of effective alleles (Ne), expected heterozygosity (HE), observed heterozygosity (HO), inbreeding coefficient (FIS), and fixation index (FST) across 24 years, differentiating by gate type, and comparing pre-(1998–2009) and post-(2010–2021) studbook cap periods using regression analysis. Results: Our results support decreased genetic diversity for both trotters and pacers over time. However, pacing Standardbreds exhibited significantly slower rates of decrease in genetic diversity after the 2009 studbook cap, as evidenced by Ne, HE, and FIS (PBonferroni < 0.01). Additionally, moderate levels of genetic differentiation were found between trotters and pacers (0.05 < FST < 0.09), which increased over time. Conclusions: Given that the rate of loss of diversity does not appear to differ pre and post studbook cap in trotters and that there is an increase in genetic differentiation between the groups over time, developing additional breeding tools and strategies is necessary to help the subpopulation mitigate further decline.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►
Show Figures
Open AccessArticle
Codon Usage Bias in Mitochondrial Genomes Across Three Species of Siphonaria (Mollusca: Gastropoda)
by
Jingjing Gu, Xuan Zhou, Chao Song, Yiyi Wang, Haobo Jin, Teng Lei and Xin Qi
Genes 2025, 16(7), 747; https://doi.org/10.3390/genes16070747 (registering DOI) - 26 Jun 2025
Abstract
Background: Siphonaria is a genus of false limpets belonging to the Gastropoda class. Only two species of this genus have been described with mitochondrial genomes. Moreover, the codon usage patterns and factors influencing them have not been studied. This study aims to expand
[...] Read more.
Background: Siphonaria is a genus of false limpets belonging to the Gastropoda class. Only two species of this genus have been described with mitochondrial genomes. Moreover, the codon usage patterns and factors influencing them have not been studied. This study aims to expand the mitochondrial genome data of this genus and clarify the codon usage patterns. Methods: The complete mitochondrial genome of Siphonaria japonica was sequenced using next-generation sequencing. The gene arrangement and phylogenetic status were compared with Siphonaria gigas and Siphonaria pectinata. The codon usage bias of the three mitochondrial genomes was analyzed based on the relative synonymous codon usage (RSCU), the effective number of codons (ENC) plot, the parity rule 2 (PR2)-bias plot, and neutrality plot analyses. Results: The gene arrangement and maximum-likelihood phylogenetic tree support a close relationship between S. japonica and S. pectinata. The codon usage bias analysis indicated that the codon usage bias of mitochondrial PCGs in the three species was primarily influenced by natural selection. Conclusions: This study offers significant evolutionary insights into the phylogenetic relationships and molecular adaptation strategies among Siphonaria species.
Full article
(This article belongs to the Section Population and Evolutionary Genetics and Genomics)
►▼
Show Figures
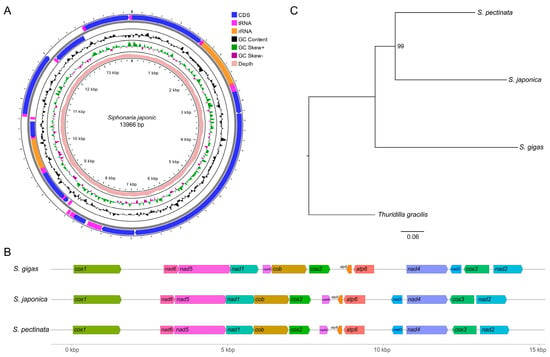
Figure 1
Open AccessArticle
Hwanhon Decoction Ameliorates Cognitive Impairment and Suppresses Neuroinflammation in a Chronic Cerebral Hypoperfusion Mouse Model: Involvement of Key Genes Identified by Network Pharmacology
by
Sieun Kang, Chiyeon Lim, Sehyun Lim, Kyoung-Min Kim and Suin Cho
Genes 2025, 16(7), 746; https://doi.org/10.3390/genes16070746 (registering DOI) - 26 Jun 2025
Abstract
Background: With an aging population, dementia prevalence is increasing in Korea. Vascular dementia (VaD), often caused by cerebrovascular disease (CVD), is more common in Korea compared to Western countries. Hwanhon decoction, a traditional medicine containing Ephedrae Herba, Armeniacae Semen, and Glycyrrhizae Radix et
[...] Read more.
Background: With an aging population, dementia prevalence is increasing in Korea. Vascular dementia (VaD), often caused by cerebrovascular disease (CVD), is more common in Korea compared to Western countries. Hwanhon decoction, a traditional medicine containing Ephedrae Herba, Armeniacae Semen, and Glycyrrhizae Radix et Rhizoma, is traditionally used for CVD-related loss of consciousness. This study aimed to assess the cognitive improvement and anti-inflammatory effects of Hwanhon decoction extract (HHex) in a mouse model of VaD caused by chronic cerebral hypoperfusion (CCH). Methods: Key pharmacologically active ingredients of Hwanhon decoction were identified using network pharmacology analysis. VaD was induced in C57Bl/6 male mice through bilateral common carotid artery stenosis (BCAS). Mice were divided into sham surgery, BCAS control, low-dose HHex (L-HHex), and high-dose HHex (H-HHex) groups (n = 5/group). After CCH induction, L-HHex or H-HHex was administered thrice weekly for six weeks. Cognitive function, inflammatory markers, and RNA sequencing data were analyzed. Results: HHex administration reduced cognitive impairment and mitigated CCH-induced astrocyte activation. Inflammatory responses mediated by reactive astrocytes were suppressed, and network pharmacology predicted central proteins influencing HHex’s activity. Conclusions: HHex alleviated cognitive dysfunction and reduced inflammation in a VaD mouse model, suggesting its potential as a therapeutic agent for vascular dementia associated with impaired cerebral blood flow.
Full article
(This article belongs to the Special Issue Genetics and Treatment in Neurodegenerative Diseases)
►▼
Show Figures
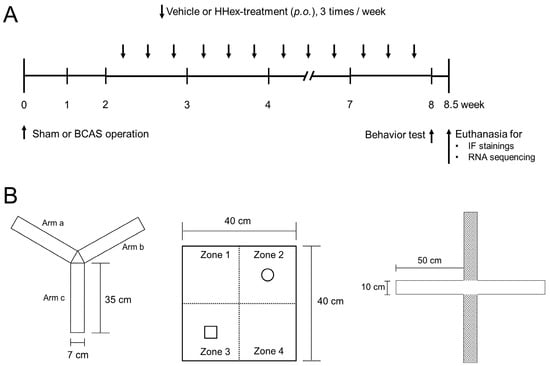
Figure 1
Open AccessArticle
Phenotypic and Genotypic Characterization of 171 Patients with Syndromic Inherited Retinal Diseases Highlights the Importance of Genetic Testing for Accurate Clinical Diagnosis
by
Sofia Kulyamzin, Rina Leibu, Hadas Newman, Miriam Ehrenberg, Nitza Goldenberg-Cohen, Shiri Zayit-Soudry, Eedy Mezer, Ygal Rotenstreich, Iris Deitch, Daan M. Panneman, Dinah Zur, Elena Chervinsky, Stavit A. Shalev, Frans P. M. Cremers, Dror Sharon, Susanne Roosing and Tamar Ben-Yosef
Genes 2025, 16(7), 745; https://doi.org/10.3390/genes16070745 (registering DOI) - 26 Jun 2025
Abstract
Background: Syndromic inherited retinal diseases (IRDs) are a clinically and genetically heterogeneous group of disorders, involving the retina and additional organs. Over 80 forms of syndromic IRD have been described. Methods: We aimed to phenotypically and genotypically characterize a cohort of 171 individuals
[...] Read more.
Background: Syndromic inherited retinal diseases (IRDs) are a clinically and genetically heterogeneous group of disorders, involving the retina and additional organs. Over 80 forms of syndromic IRD have been described. Methods: We aimed to phenotypically and genotypically characterize a cohort of 171 individuals from 140 Israeli families with syndromic IRD. Ophthalmic examination included best corrected visual acuity, fundus examination, visual field testing, retinal imaging and electrophysiological evaluation. Most participants were also evaluated by specialists in fields relevant to their extra-retinal symptoms. Genetic analyses included haplotype analysis, homozygosity mapping, Sanger sequencing and next-generation sequencing. Results: In total, 51% of the families in the cohort were consanguineous. The largest ethnic group was Muslim Arabs. The most common phenotype was Usher syndrome (USH). The most common causative gene was USH2A. In 29% of the families, genetic analysis led to a revised or modified clinical diagnosis. This included confirmation of an atypical USH diagnosis for individuals with late-onset retinitis pigmentosa (RP) and/or hearing loss (HL); diagnosis of Heimler syndrome in individuals with biallelic pathogenic variants in PEX6 and an original diagnosis of USH or nonsyndromic RP; and diagnosis of a mild form of Leber congenital amaurosis with early-onset deafness (LCAEOD) in an individual with a heterozygous pathogenic variant in TUBB4B and an original diagnosis of USH. Novel genotype–phenotype correlations included biallelic pathogenic variants in KATNIP, previously associated with Joubert syndrome (JBTS), in an individual who presented with kidney disease and IRD, but no other features of JBTS. Conclusions: Syndromic IRDs are a highly heterogeneous group of disorders. The rarity of some of these syndromes on one hand, and the co-occurrence of several syndromic and nonsyndromic conditions in some individuals, on the other hand, complicates the diagnostic process. Genetic analysis is the ultimate way to obtain an accurate clinical diagnosis in these individuals.
Full article
(This article belongs to the Special Issue Advances in Medical Genetics)
►▼
Show Figures
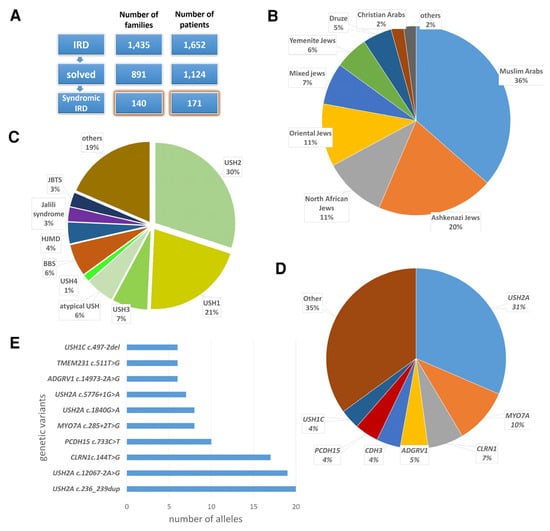
Figure 1
Open AccessArticle
Transcriptomic Insights into GABA Accumulation in Tomato via CRISPR/Cas9-Based Editing of SlGAD2 and SlGAD3
by
Jin-Young Kim, Yu-Jin Jung, Dong Hyun Kim and Kwon-Kyoo Kang
Genes 2025, 16(7), 744; https://doi.org/10.3390/genes16070744 (registering DOI) - 26 Jun 2025
Abstract
Background: γ-Aminobutyric acid (GABA) is a non-proteinogenic amino acid with key roles in plant metabolism, stress responses, and fruit nutritional quality. In tomato (Solanum lycopersicum), GABA levels are dynamically regulated during fruit development but decline in the late ripening stages.
[...] Read more.
Background: γ-Aminobutyric acid (GABA) is a non-proteinogenic amino acid with key roles in plant metabolism, stress responses, and fruit nutritional quality. In tomato (Solanum lycopersicum), GABA levels are dynamically regulated during fruit development but decline in the late ripening stages. Methods: To enhance GABA accumulation, we used CRISPR/Cas9 to edit the calmodulin-binding domain (CaMBD) of SlGAD2 and SlGAD3, which encode glutamate decarboxylases (GADs). The resulting truncated enzymes were expected to be constitutively active. We quantified GABA content in leaves and fruits and performed transcriptomic analysis on edited lines at the BR+7 fruit stage. Results: CaMBD truncation significantly increased GABA levels in both leaves and fruits. In gad2 sg1 lines, GABA levels increased by 3.5-fold in leaves and 3.2-fold in BR+10 fruits; in gad3 sg3 lines, increases of 2.8- and 2.5-fold were observed, respectively. RNA-seq analysis identified 1383 DEGs in gad2 #1−5 and 808 DEGs in gad3 #3−8, with 434 DEGs shared across both lines. These shared DEGs showed upregulation of GAD, GABA-T, and SSADH, and downregulation of stress-responsive transcription factors including WRKY46, ERF, and NAC. Notably, total free amino acid content and fruit morphology remained unchanged despite elevated GABA. Conclusions: CRISPR/Cas9-mediated editing of the CaMBD in SlGAD genes selectively enhances GABA biosynthesis in tomato without adverse effects on development or fruit quality. These lines offer a useful platform for GABA-centered metabolic engineering and provide insights into GABA’s role in transcriptional regulation during ripening.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
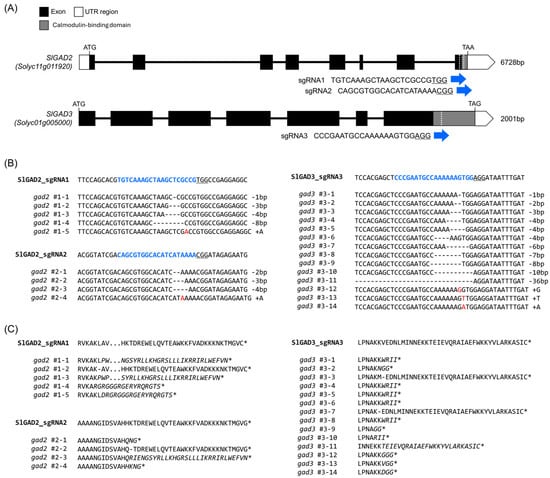
Figure 1
Open AccessArticle
Exploring Concomitant Ophthalmic Comorbidities in Portuguese Patients with Inherited Retinal Diseases: A Comprehensive Clinical Study
by
Rita Mesquita, Ana Marta, Pedro Marques-Couto, José Costa, Sérgio Estrela-Silva, Diogo Cabral, João Pedro Marques and Sara Vaz-Pereira
Genes 2025, 16(7), 743; https://doi.org/10.3390/genes16070743 (registering DOI) - 26 Jun 2025
Abstract
Background/Objectives: Inherited retinal diseases (IRDs) are a heterogeneous group of rare eye disorders characterized by progressive photoreceptor degeneration, leading to severe visual impairment or even blindness. This study aims to investigate the prevalence, types, and clinical significance of ophthalmic comorbidities in Portuguese
[...] Read more.
Background/Objectives: Inherited retinal diseases (IRDs) are a heterogeneous group of rare eye disorders characterized by progressive photoreceptor degeneration, leading to severe visual impairment or even blindness. This study aims to investigate the prevalence, types, and clinical significance of ophthalmic comorbidities in Portuguese patients with IRDs. Methods: This nationwide Portuguese population-based retrospective study was based on the IRD-PT registry (retina.com.pt). Statistical analysis was conducted using Microsoft® Excel® for Microsoft 365 and IBM SPSS Statistics version 29.0.2.0. Informed consent was obtained from all participants. Results: A total of 1531 patients (1254 families) from six centers were enrolled. The cohort consisted of 51% males, with a mean age of 45.8 ± 19.3 years and a mean age at diagnosis of 39.4 ± 19.5 years. Overall, ocular comorbidities were reported in 644 patients (42.1%). In 176 individuals (11.5%), multiple concurrent comorbidities were found. Cataract was the most common comorbidity (21.3%), followed by amblyopia (6.3%) and high myopia (5.9%). Statistically significant associations with ocular comorbidities were observed in isolated progressive IRDs. Specifically, AR RP was associated with cataract (p < 0.001), and gene analysis revealed several significant associations. CRB1 was statistically linked to epiretinal membrane (ERM) (p = 0.003), EYS with cataract (p = 0.001), PROM1 with choroidal neovascularization (CNV) (p = 0.0026), and USH2A with macular hole (p = 0.01). Patients with the RPE65 mutation in Leber congenital amaurosis were associated with ERM (p = 0.019). There was also a significant association between X-linked RP and high myopia (p < 0.001) and CNV in Best disease (p < 0.001); in syndromic IRDs, cataract, cystoid macular edema, and ERM were observed in Usher syndrome, p = 0.002, p = 0.002, and p = 0.005, respectively, and the MYO7A gene was linked to cataract (p = 0.041) and strabismus (p = 0.013); pseudoxanthoma elasticum was significantly associated with CNV (p = 0.002); and foveal hypoplasia was associated with anterior segment dysgenesis (p < 0.001). Conclusions: This study enhances the current understanding of ocular comorbidities in IRDs in Portuguese patients. Common findings were cataract, refractive error, and CME. Stationary IRDs and pattern dystrophies showed fewer concomitant comorbidities, supporting their classification as non-progressive or benign conditions. The significance of registries like IRD-PT cannot be overstated, particularly in the context of rare diseases. These databases serve multiple crucial functions in enabling detailed documentation of disease characteristics and long-term monitoring of disease progression.
Full article
(This article belongs to the Special Issue Genetics in Retinal Diseases—2nd Edition)
►▼
Show Figures
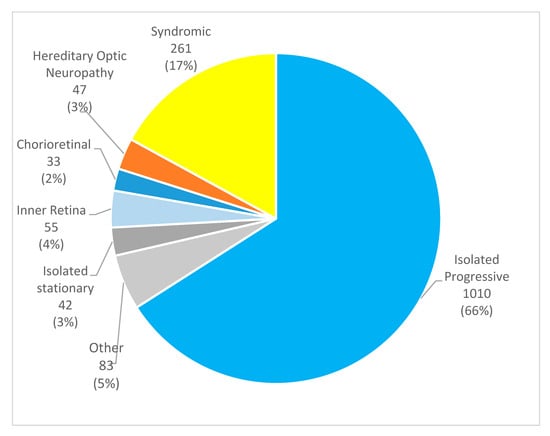
Figure 1
Open AccessArticle
Identification and Bioinformatics Analysis of the HSP20 Family in the Peony
by
Haoran Ma, Heling Yuan, Wenxuan Bu, Minhuan Zhang, Yu Huang, Jian Hu and Jiwu Cao
Genes 2025, 16(7), 742; https://doi.org/10.3390/genes16070742 (registering DOI) - 26 Jun 2025
Abstract
Background: The peony (Paeonia suffruticosa Andr.), a globally valued woody ornamental species, suffers severe heat-induced floral damage that compromises its horticultural value. While the HSP20 proteins are critical for plant thermotolerance, their genomic organization and regulatory dynamics remain uncharacterized in the peony.
[...] Read more.
Background: The peony (Paeonia suffruticosa Andr.), a globally valued woody ornamental species, suffers severe heat-induced floral damage that compromises its horticultural value. While the HSP20 proteins are critical for plant thermotolerance, their genomic organization and regulatory dynamics remain uncharacterized in the peony. This study aims to systematically identify the PsHSP20 genes, resolve their molecular features, and elucidate their heat-responsive expression patterns to enable targeted thermotolerance breeding. Methods: The genome-wide identification employed HMMER and BLASTP searches against the peony genome. The physicochemical properties and protein structures of the gene family were analyzed using online websites, such as Expasy, Plant-mPLoc, and SOPMA. The cis-regulatory elements were predicted using PlantCARE. Expression profiles under different times of 40 °C heat stress were validated by qRT-PCR (p < 0.05). Results: We identified 58 PsHSP20 genes, classified into 11 subfamilies. All members retain the conserved α-crystallin domain, and exhibit predominant nuclear/cytoplasmic localization. Chromosomal mapping revealed uneven distribution without lineage-specific duplications. The promoters were enriched in stress-responsive elements (e.g., HSE, ABRE) and in 24 TF families. The protein networks linked 13 PsHSP20s to co-expressed partners in heat response (GO:0009408) and ER protein processing (KEGG:04141). Transcriptomics demonstrated rapid upregulation of 48 PsHSP20s within 2 h of heat exposure, with PsHSP20-12, -34, and -51 showing the highest induction (>15-fold) at 6 h/24 h. Conclusions: This first genome-wide study resolves the architecture and heat-responsive dynamics of the PsHSP20 family. The discovery of early-induced genes (PsHSP20-12/-34/-51) provides candidates for thermotolerance enhancement. These findings establish a foundation for molecular breeding in the peony.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
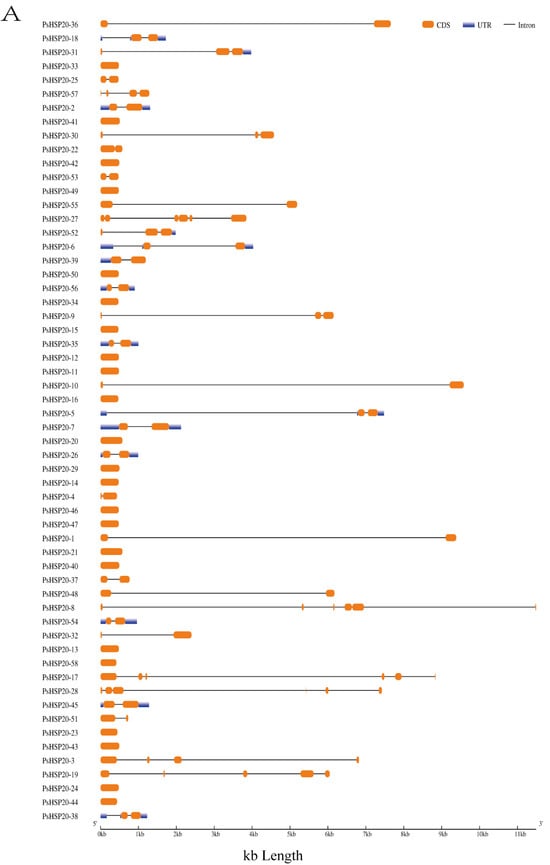
Figure 1
Open AccessArticle
Genomic Regions Associated with Respiratory Disease in Holstein Calves in the Southern United States
by
Allison L. Herrick, Jennifer N. Kiser, Stephen N. White and Holly L. Neibergs
Genes 2025, 16(7), 741; https://doi.org/10.3390/genes16070741 (registering DOI) - 26 Jun 2025
Abstract
Background/Objectives: Bovine respiratory disease (BRD) is a common disease impacting cattle throughout the US. BRD is a multifactorial disease as disease risk varies with the genetic profile of the host, environmental conditions, and pathogen exposure. Selection for enhanced BRD resistant cattle can aid
[...] Read more.
Background/Objectives: Bovine respiratory disease (BRD) is a common disease impacting cattle throughout the US. BRD is a multifactorial disease as disease risk varies with the genetic profile of the host, environmental conditions, and pathogen exposure. Selection for enhanced BRD resistant cattle can aid in reducing BRD. The objectives of this study were to identify loci, gene sets, and genes associated and enriched for BRD in pre- and post-weaned Holstein cattle. Methods: Cases consisted of 2147 and 5607 calves treated for BRD as pre-weaned (0–60 days old) and post-weaned (61–420 days old) calves, respectively. Controls consisted of calves untreated for BRD that remained in the herd for 61 (n = 14,219) days for pre-weaned or 421 (n = 12,242) days for post-weaned calves. A genome-wide association analysis (GWAA) identified loci and positional candidate genes associated with BRD (uncorrected P < 1 × 10−5) for additive, dominant, and recessive inheritance models. A gene set enrichment analysis (GSEA-SNP) identified gene sets and leading-edge genes enriched (NES ≥ 3) for BRD. Results: In pre-weaned calves, 62 loci and 123 positional candidate genes were associated (P < 1 × 10−5) in addition to the 12 gene sets and 126 leading-edge genes enriched (NES ≥ 3) for BRD. In post-weaned calves, 181 loci and 185 positional candidate genes were associated (P < 1 × 10−5), and 63 gene sets and 849 leading-edge genes were enriched (NES ≥ 3) for BRD. Conclusions: These results provide further insight and validation of genomic regions that enhance selection for BRD resistance and for healthier cattle.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
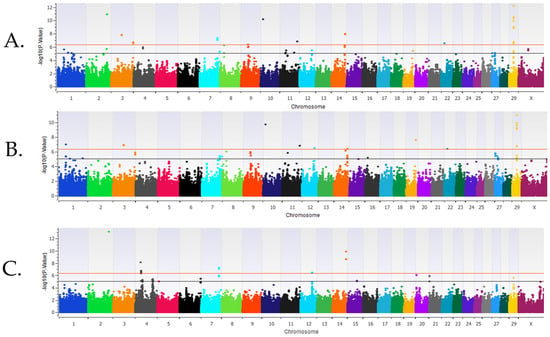
Figure 1
Open AccessArticle
Characterizing Gene-Level Adaptations in the Gut Microbiome During Viral Infections: The Role of a Fucoidan-Rich Extract
by
Gissel García, Josanne Soto, Carmen Valenzuela and Raul De Jesús Cano
Genes 2025, 16(7), 740; https://doi.org/10.3390/genes16070740 (registering DOI) - 26 Jun 2025
Abstract
Background/Objectives: This study aimed to examine the effects of a Fucoidan-rich extract from Saccharina latissima (SLE-F) on differential gut microbiota composition, intestinal inflammation status, and microbial functional gene expression in participants infected with Dengue or Oropouche virus at the Hermanos Ameijeiras Hospital in
[...] Read more.
Background/Objectives: This study aimed to examine the effects of a Fucoidan-rich extract from Saccharina latissima (SLE-F) on differential gut microbiota composition, intestinal inflammation status, and microbial functional gene expression in participants infected with Dengue or Oropouche virus at the Hermanos Ameijeiras Hospital in Havana, Cuba. Methods: Fecal samples were collected at baseline, day 28, and day 90 from 90 healthy adults, some of whom contracted the virus during the study period. Functional gene analysis was conducted using two approaches—the Kruskal–Wallis H test and linear discriminant analysis effect size—applied to ortholog-level data normalized by read count and gene copy number. Results: Infected participants exhibited significantly lower Lachnospiraceae-to-Enterobacteriaceae (LE) ratios, indicating increased intestinal inflammation. High-dose SLE-F treatment led to a significant reduction in the LE ratio (p = 0.006), suggesting a strong anti-inflammatory effect. Microbiome analysis revealed a shift from dysbiosis to a more balanced composition by the end of the study, characterized by increased abundances of Akkermansia muciniphila, Bifidobacterium adolescentis, and B. longum, along with decreased pro-inflammatory taxa such as Fusobacterium. Conclusions: Genetic analysis provided distinct yet complementary insights into the microbiome’s functional responses to infection and therapeutic modulation by Fucoidan. These findings highlight the therapeutic potential of high-dose Fucoidan in reducing gut inflammation and promoting microbiome recovery following viral infections.
Full article
(This article belongs to the Section Microbial Genetics and Genomics)
►▼
Show Figures
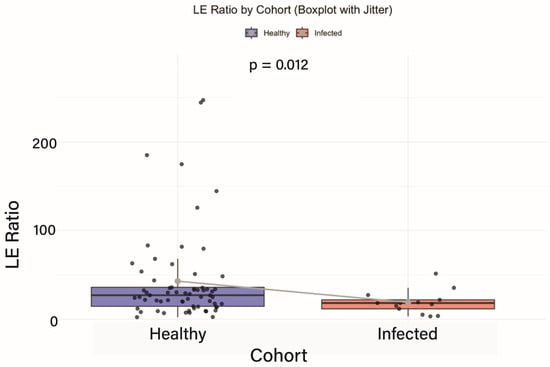
Figure 1
Open AccessArticle
Genome Annotation and Catalytic Profile of Rhodococcus rhodochrous IEGM 107, Mono- and Diterpenoid Biotransformer
by
Natalia A. Plotnitskaya, Polina Yu. Maltseva and Irina B. Ivshina
Genes 2025, 16(7), 739; https://doi.org/10.3390/genes16070739 (registering DOI) - 26 Jun 2025
Abstract
Background/Objectives: Rhodococcus rhodochrous IEGM 107 cells exhibit pronounced catalytic activity toward mono- and diterpenoids. However, the genetics and enzymatic foundations underlying this activity remain poorly understood. Methods: Using new-generation sequencing, the R. rhodochrous IEGM 107 whole genome was sequenced. Bioinformatic analysis
[...] Read more.
Background/Objectives: Rhodococcus rhodochrous IEGM 107 cells exhibit pronounced catalytic activity toward mono- and diterpenoids. However, the genetics and enzymatic foundations underlying this activity remain poorly understood. Methods: Using new-generation sequencing, the R. rhodochrous IEGM 107 whole genome was sequenced. Bioinformatic analysis and PCR were employed to identify and characterize genes, with a focus on cytochromes P450 (CYP450s). Results: The catalytic potential of R rhodochrous IEGM 107 was revealed. Its CYP450 genes were detected and analyzed, providing information on the enzymatic base of the strain related to the biotransformation of terpenoids. Conclusions: These findings enhance the understanding of the molecular and genetic basis for terpenoid transformations in R. rhodochrous actinomycetes. The results provide a foundation for future studies on gene expression and enzyme characterization aimed at developing efficient and selective biocatalysts for mono- and diterpenoid transformations.
Full article
(This article belongs to the Section Microbial Genetics and Genomics)
►▼
Show Figures
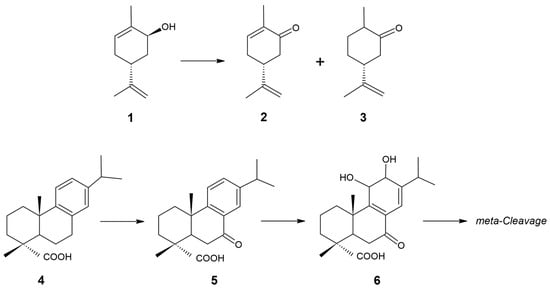
Figure 1
Open AccessArticle
Endplate Lesions of the Lumbar Spine: Biochemistry and Genetics
by
Alessandra Colombini, Vincenzo Raffo, Angela Elvira Covone, Tito Bassani, Domenico Coviello, Sabina Cauci, Ludovica Pallotta and Marco Brayda-Bruno
Genes 2025, 16(7), 738; https://doi.org/10.3390/genes16070738 (registering DOI) - 26 Jun 2025
Abstract
Background/Objectives: Endplate lesions of the lumbar spine are often asymptomatic and frequently observed incidentally by radiological assessment. Variants in the vitamin D receptor gene (VDR) and an increase in some biochemical markers related to the osteo-cartilaginous metabolism were found in patients
[...] Read more.
Background/Objectives: Endplate lesions of the lumbar spine are often asymptomatic and frequently observed incidentally by radiological assessment. Variants in the vitamin D receptor gene (VDR) and an increase in some biochemical markers related to the osteo-cartilaginous metabolism were found in patients with endplate lesions. The aim of this study was to identify biochemical and genetic markers putatively associated with the presence of endplate lesions of the lumbar spine. Methods: Quantification of circulating bone remodeling proteins was obtained from 10 patients with endplate lesions and compared with age- and sex-matched controls. Whole exome sequencing (WES) was performed on patient genomic DNA using the Novaseq 6000 platform (Illumina, San Diego, CA, USA), obtaining a median read depth of 117×–200×, with ≥98% of regions covering at least 20×. The sequencing product was aligned to the reference genome (GRCh38.p13-hg38) and analyzed with Geneyx software. Results: We observed modifications in the levels of circulating proteins involved in bone remodeling and angiogenesis. We identified variants of interest in aggrecan (ACAN), bone morphogenetic protein 4 (BMP4), cytochrome P450 family 3 subfamily A member 4 (CYP3A4), GLI family zinc finger 2 (GLI2), heparan sulfate proteoglycan 2 (HSPG2), and mesoderm posterior bHLH transcription factor 2 (MESP2). VDR polymorphism (rs2228570) was present in nine patients, with the homozygotic ones having more severe endplate lesions and higher levels of the analyzed circulating markers in comparison with heterozygotic patients. Conclusions: These data represent interesting evidence of genetic variants, particularly in VDR, and altered levels of circulating markers of bone remodeling associated with endplate lesions, which should be confirmed in a larger population. The hypothesis suggested by our results is that the endplate lesions could be the consequence of an altered ossification mechanism at the vertebral level.
Full article
(This article belongs to the Special Issue Genes and Gene Polymorphisms Associated with Complex Diseases)
►▼
Show Figures
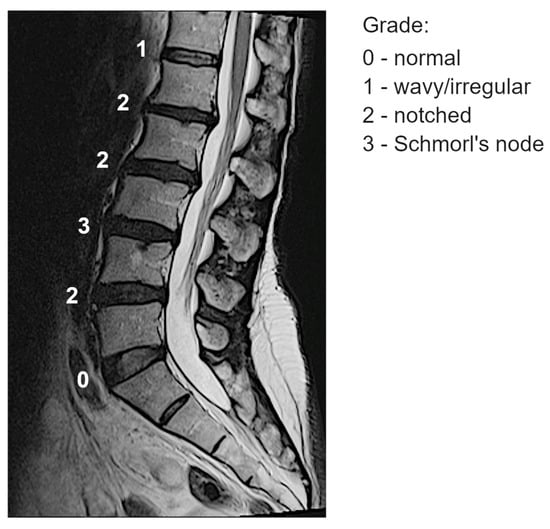
Figure 1
Open AccessArticle
MYB Confers Sorafenib Resistance in Human Leukemia Cells via Inhibiting Ferroptosis Through FTH1 Upregulation
by
Xiaoxiao Tao, Yucheng Wang, Siyu Shen, Huiying Fang, Hongkuan Song, Junfang Zhang and Bingshe Han
Genes 2025, 16(7), 737; https://doi.org/10.3390/genes16070737 (registering DOI) - 26 Jun 2025
Abstract
Background: MYB is a key transcription factor that plays an essential role in regulating hematopoiesis, particularly influencing cell proliferation, differentiation, and apoptosis. It has been extensively implicated in the pathogenesis and progression of leukemia, as well as in determining patient prognosis and responsiveness
[...] Read more.
Background: MYB is a key transcription factor that plays an essential role in regulating hematopoiesis, particularly influencing cell proliferation, differentiation, and apoptosis. It has been extensively implicated in the pathogenesis and progression of leukemia, as well as in determining patient prognosis and responsiveness to chemotherapy. Despite these well-documented roles, the precise molecular mechanisms by which MYB contributes to chemotherapy resistance in leukemia remain largely undefined. Methods: In this study, we investigated the potential role of MYB in regulating ferroptosis, a form of regulated cell death driven by iron-dependent lipid peroxidation, which has recently emerged as a novel therapeutic target in cancer. We overexpressed and knockdown MYB in human leukemia K562 cells and evaluated changes in ferroptosis-related markers, as well as cell proliferation and migration capacities, in the context of treatment with the chemotherapeutic agent sorafenib. Results: Our findings demonstrated that MYB overexpression significantly enhanced the resistance of human leukemia cells to sorafenib, while MYB knockdown increased their drug sensitivity. Mechanistically, MYB was found to upregulate ferritin heavy chain 1 (FTH1), thereby suppressing sorafenib-induced ferroptosis and cell death. Further, FTH1 knockdown significantly reduced the proliferation and migration ability of K562 cells and enhanced sorafenib-induced ferroptosis. Rescue experiments confirmed that FTH1 is required for MYB induced sorafenib resistance and ferroptosis inhibition in human leukemia cells. Conclusions: Collectively, this study identifies the MYB-FTH1 axis as a novel regulatory pathway modulating ferroptosis and chemoresistance in leukemia cells, providing potential therapeutic targets for improving treatment precision and preventing disease relapse.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures
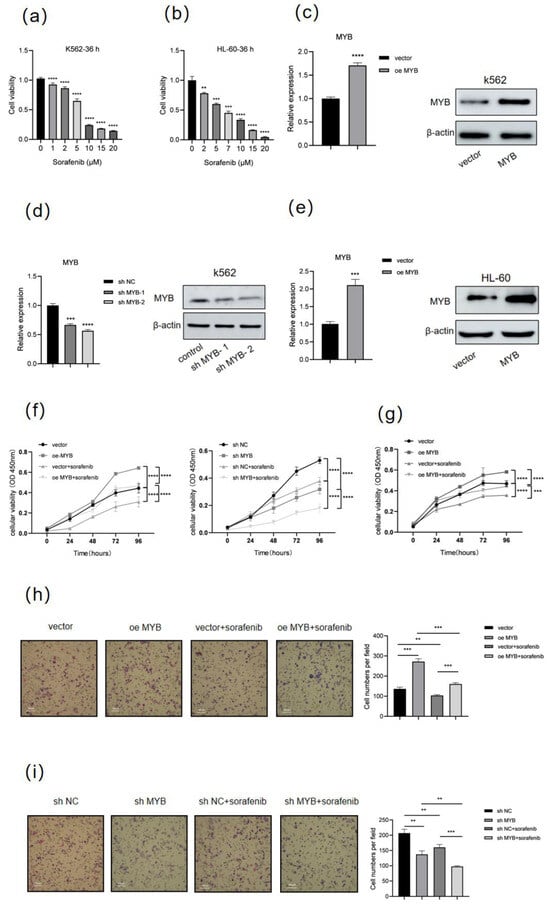
Figure 1
Open AccessArticle
Characterization of Constitutional Ring Chromosomes over 37 Years of Experience at a Single-Site Institution
by
Jaclyn B. Murry and Barbara R. DuPont
Genes 2025, 16(7), 736; https://doi.org/10.3390/genes16070736 (registering DOI) - 25 Jun 2025
Abstract
Background/Objectives: Ring chromosomes (RCs) can be rare or common depending on the chromosome involved. With interest in the historical RCs identified by our laboratory, we curated instances to provide further information to this research field. Methodology: We carried out a retrospective, single-center
[...] Read more.
Background/Objectives: Ring chromosomes (RCs) can be rare or common depending on the chromosome involved. With interest in the historical RCs identified by our laboratory, we curated instances to provide further information to this research field. Methodology: We carried out a retrospective, single-center study of constitutional RCs identified starting in the late 1980s. Details for 40 RCs with a modal number of 46 chromosomes are featured here. Results: Mosaic and non-mosaic RCs are identified, with a preponderance of pediatric-aged females at first ascertainment. We corroborated an enrichment for acrocentric and X chromosome RCs. Six were ascertained perinatally, with peripheral blood being the most commonly studied postnatal specimen type. Notable RCs included a female fetus with an increased risk for monosomy X, whose mosaic RCY arose secondary to a translocation between the sex chromosomes. In another, a series of complex events formed three structurally aberrant chromosomes, including an RC4 with loss of 4p16.3, corresponding with the observed phenotypic expression of Wolf–Hirschhorn syndrome. In another, a mosaic RCX was co-identified with an isochromosome 21q, resulting in a dual diagnosis of trisomy 21 and Turner syndrome. In another, the atypical RC21 structure raises the possibility of a complex rearrangement. Chromosomal microarray data clarified breakpoints and gene dosage imbalances in fifteen, while low-level mosaicism for the RC escaped detection by array in another. Eight RCs were de novo, and parental exclusion was documented for six. Conclusions: This study illustrates the need for cytogenomic follow-up to improve the literature for patients with RCs.
Full article
(This article belongs to the Special Issue Clinical Cytogenetics: Current Advances and Future Perspectives)
►▼
Show Figures
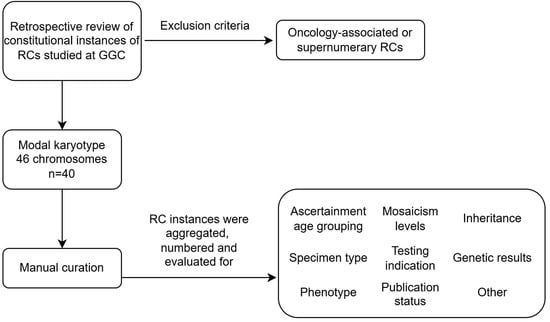
Figure 1
Open AccessBrief Report
Functional and Regulatory Effects of Factor V Leiden and Factor V rs6028 in Breast Cancer
by
Sara Marie Lind, Marit Sletten, Carola Elisabeth Henriksson, Mari Tinholt and Nina Iversen
Genes 2025, 16(7), 735; https://doi.org/10.3390/genes16070735 (registering DOI) - 25 Jun 2025
Abstract
Background/Objectives: Cancer progression and the hemostatic system are closely linked. Coagulation factor V (FV) has a key function in coagulation, with both pro- and anticoagulant functions. FV gene (F5) expression and F5 variants have been linked to breast cancer progression.
[...] Read more.
Background/Objectives: Cancer progression and the hemostatic system are closely linked. Coagulation factor V (FV) has a key function in coagulation, with both pro- and anticoagulant functions. FV gene (F5) expression and F5 variants have been linked to breast cancer progression. The direct impact of F5 variants on FV expression and functional effects in breast cancer are unknown. We aimed to investigate whether the F5 variants FV Leiden (F5 rs6025) and F5 rs6028 influenced FV expression, coagulant activity, and apoptosis in breast cancer cells. Methods: MDA-MB-231 cells were transfected with overexpression plasmids containing F5 wild type, F5 rs6025 or F5 rs6028. We investigated the functional impact of the F5 variants on F5 mRNA, FV protein, FV coagulant activity, and apoptosis in vitro, and examined the potential of the variants as transcriptional regulators of F5 expression in silico. Results: Increased F5 mRNA, FV protein, and apoptosis were observed in cells transfected with F5 wild-type overexpression plasmid compared to empty vector. F5 mRNA, protein, coagulant activity, and apoptosis were further increased with the F5 rs6025 and F5 rs6028 variants compared to F5 wild type. Cis-expression quantitative trait loci analyses indicated a regulatory effect of F5 rs6028, and putative transcription factor binding sites for several transcription factors overlapped with the position of F5 rs6025. Conclusions: Our study demonstrated that F5 rs6025 and F5 rs6028 have a regulatory effect on FV synthesis that might affect apoptosis in breast cancer. The F5 variants might therefore enhance the tumor suppressor function of FV in breast cancer.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures
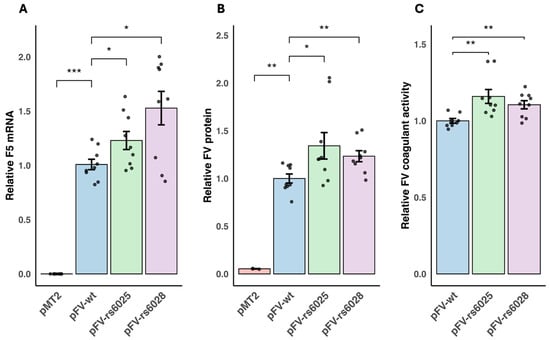
Figure 1
Open AccessArticle
Genome-Wide Identification and Analysis of bZIP Transcription Factor Gene Family in Broomcorn Millet (Panicum miliaceum L.)
by
Peipei An, Tianxiang Liu, Zhijie Shui, Panrong Ren and Shan Duan
Genes 2025, 16(7), 734; https://doi.org/10.3390/genes16070734 - 24 Jun 2025
Abstract
Background: Basic (region) leucine zippers (bZIPs) make up one of the largest families and are some of the most prevalent evolutionarily conserved transcription factors (TFs) in eukaryotic organisms. Plant bZIP family members are involved in seed germination, vegetative growth, flower development, light response,
[...] Read more.
Background: Basic (region) leucine zippers (bZIPs) make up one of the largest families and are some of the most prevalent evolutionarily conserved transcription factors (TFs) in eukaryotic organisms. Plant bZIP family members are involved in seed germination, vegetative growth, flower development, light response, and various biotic/abiotic stress response pathways. Nevertheless, a detailed identification and genome-wide analysis of the bZIP family genes in broomcorn millet have not been conducted. Methods: In this research, we performed genome-wide identification, phylogenetic analysis, cis-elements analysis, and expression pattern analysis. Results: 144 bZIP transcription factors were identified from the P. miliaceum genome and classified into eleven subfamilies using a phylogenetic analysis. Motif and bZIP domain sequence alignment analyses indicated that the members in each subfamily were relatively conserved. Furthermore, a promoter analysis revealed that bZIP transcription factor family genes were responsive to multiple hormones and environmental stresses. Additionally, cis-element MYB binding sites were identified in the promoters of most PmbZIP genes. A gene expression analysis showed that 18 PmbZIP genes were differentially expressed during seed germination in salt stress, with 7 being significantly downregulated and 11 upregulated, thus suggesting that these PmbZIP genes may play an important role in the salt stress response and seed germination. Conclusions: Current research provides valuable information for further functional analyses of the PmbZIP gene family and as a reference for future studies on broomcorn millet’s stress response.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
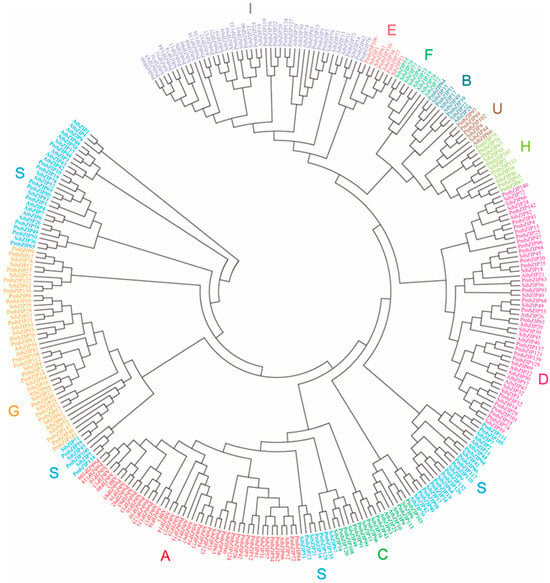
Figure 1
Open AccessArticle
Genome-Wide Associations with Body and Fleece Weight in United States Sheep
by
Gabrielle M. Becker, Daniel Schaub, J. Bret Taylor, Michelle R. Mousel, Carrie S. Wilson, Jamin A. Smitchger, Jacob W. Thorne and Brenda M. Murdoch
Genes 2025, 16(7), 733; https://doi.org/10.3390/genes16070733 - 24 Jun 2025
Abstract
Background/Objectives: Wool is an important product in sheep production, but the genetic mechanisms underpinning variation in wool growth are not fully understood. Identifying the genes and genomic variants that play a role in increasing fleece weight may allow for increased selection accuracy
[...] Read more.
Background/Objectives: Wool is an important product in sheep production, but the genetic mechanisms underpinning variation in wool growth are not fully understood. Identifying the genes and genomic variants that play a role in increasing fleece weight may allow for increased selection accuracy and improved economic return to producers. Methods: A genome-wide association study (GWAS) was conducted to investigate genetic associations with lifetime fleece weight, average fleece weight and average post-lambing ewe weight for Rambouillet, Polypay, Suffolk and Columbia ewes (N = 1125). Weir–Cockerham FST and runs of homozygosity (ROH) analyses were conducted to improve detection of putative wool-related signatures. Results: Twenty-four SNPs were identified through GWAS for lifetime fleece weight, average fleece weight and average post-lambing ewe weight. Chromosomes 2 and 6 contained ROH islands in Rambouillet, and chromosomes 2, 3 and 10 contained ROH islands in Suffolk. The FST analysis identified 18 SNPs in proximity to 37 genes of interest. Conclusions: Many of the SNPs and signatures of selection reported in this study are near or within current candidate genes for wool production and wool quality, including ADAR, KCNN3, NTN1, SETBP1, TP53 and TNFSF12. The significant SNPs implicated by GWAS may be used to predict ewes’ potential for lifetime wool production and are suggested as candidates for further study to continue to elucidate the genetic mechanisms underlying wool production traits in United States sheep breeds.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
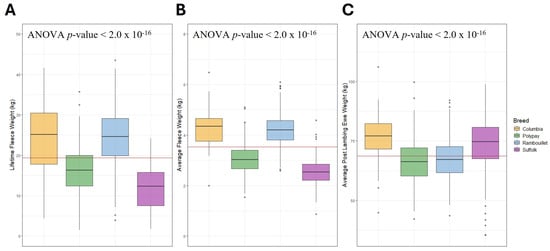
Figure 1
Open AccessReview
Radiogenomics of Stereotactic Radiotherapy: Genetic Mechanisms Underlying Radiosensitivity, Resistance, and Immune Response
by
Damir Vučinić, Ana-Marija Bukovica Petrc, Ivona Antončić, Maja Kolak Radojčić, Matea Lekić and Felipe Couñago
Genes 2025, 16(7), 732; https://doi.org/10.3390/genes16070732 - 24 Jun 2025
Abstract
Stereotactic body radiotherapy (SBRT) delivers ablative radiation doses with sub-millimeter precision. Radiogenomic studies, meanwhile, provide insights into how tumor-intrinsic genetic factors influence responses to such high-dose treatments. This review explores the radiobiological mechanisms underpinning SBRT efficacy, emphasizing the roles of DNA damage response
[...] Read more.
Stereotactic body radiotherapy (SBRT) delivers ablative radiation doses with sub-millimeter precision. Radiogenomic studies, meanwhile, provide insights into how tumor-intrinsic genetic factors influence responses to such high-dose treatments. This review explores the radiobiological mechanisms underpinning SBRT efficacy, emphasizing the roles of DNA damage response (DDR) pathways, tumor suppressor gene alterations, and inflammatory signaling in shaping tumor radiosensitivity or resistance. SBRT induces complex DNA double-strand breaks (DSBs) that robustly activate DDR signaling cascades, particularly via the ATM and ATR kinases. Tumors with proficient DNA repair capabilities often resist SBRT, whereas deficiencies in key repair genes can render them more susceptible to radiation-induced cytotoxicity. Mutations in tumor suppressor genes may impair p53-dependent apoptosis and disrupt cell cycle checkpoints, allowing malignant cells to evade radiation-induced cell death. Furthermore, SBRT provokes the release of pro-inflammatory cytokines and activates innate immune pathways, potentially leading to immunogenic cell death and reshaping the tumor microenvironment. Radiogenomic profiling has identified genomic alterations and molecular signatures associated with differential responses to SBRT and immune activation. These insights open avenues for precision radiotherapy approaches, including the use of genomic biomarkers for patient selection, the integration of SBRT with DDR inhibitors or immunotherapies, and the customization of treatment plans based on individual tumor genotypes and immune landscapes. Ultimately, these strategies aim to enhance SBRT efficacy and improve clinical outcomes through biologically tailored treatment. This review provides a comprehensive summary of current knowledge on the genetic determinants of response to stereotactic radiotherapy and discusses their implications for personalized cancer treatment.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures
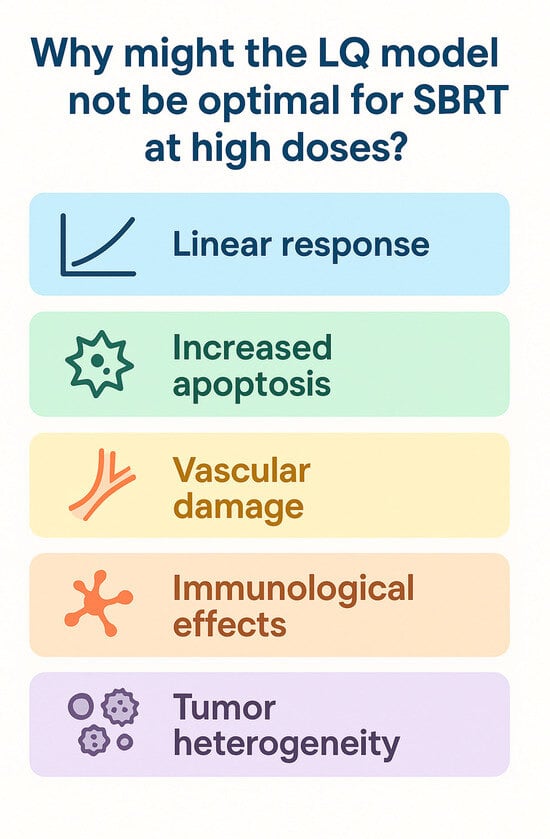
Figure 1
Open AccessArticle
Expression Dynamics and Estrogen Response of Estrogen Receptors in Duolang Sheep During Puberty
by
Lexiao Zhu, Gul Muhammad Shahbaz, Huiping Sun, Jihu Zhang, Wei Li, Ruohuai Gu and Feng Xing
Genes 2025, 16(7), 731; https://doi.org/10.3390/genes16070731 - 24 Jun 2025
Abstract
Background/Objectives: Puberty is a critical stage in sheep development when reproductive capability is established, but the hormonal mechanisms underlying this transition remain incompletely understood. This study aimed to investigate the dynamic changes in estradiol (E2) levels and the expression patterns of estrogen receptors
[...] Read more.
Background/Objectives: Puberty is a critical stage in sheep development when reproductive capability is established, but the hormonal mechanisms underlying this transition remain incompletely understood. This study aimed to investigate the dynamic changes in estradiol (E2) levels and the expression patterns of estrogen receptors (ERα and ERβ) during puberty in Duolang sheep, a breed characterized by early sexual maturity and high reproductive efficiency. Methods: A total of 18 female Duolang sheep were assigned to three developmental stages (n = 6 per group): prepuberty (145 days), puberty (within 0 h of first estrus), and postpuberty (+3 days). Serum E2 concentrations and the mRNA and protein levels of ERα and ERβ were assessed in the hypothalamus, pituitary, and ovary. Additionally, primary ovarian granulosa cells (GCs) were isolated and stimulated in vitro with increasing concentrations of E2 (0–1000 ng/mL) to evaluate the dose-dependent expression of ERα, ERβ, and gonadotropin-releasing hormone (GnRH). Results: E2 levels peaked at the onset of puberty and declined thereafter. ERα expression in the hypothalamus and pituitary decreased during puberty but rebounded postpuberty, indicating a role in negative feedback regulation. In contrast, ovarian ERα expression reached its highest level during puberty, while ERβ expression in the ovary gradually increased from prepuberty to postpuberty. In GCs, ERα exhibited a biphasic expression pattern, peaking at 250 ng/mL E2 and decreasing at higher concentrations. ERβ and GnRH expression levels increased in a dose-dependent manner. Conclusions: These findings suggest that ERα primarily mediates E2 feedback within the hypothalamus–pituitary axis, whereas ERβ is associated with ovarian development and may regulate GnRH expression during the pubertal transition. The study provides new insights into the hormonal regulation of puberty in Duolang sheep and offers potential biomarkers for improving reproductive efficiency through targeted breeding strategies.
Full article
(This article belongs to the Special Issue Gene Regulation of Development and Evolution in Mammals)
►▼
Show Figures
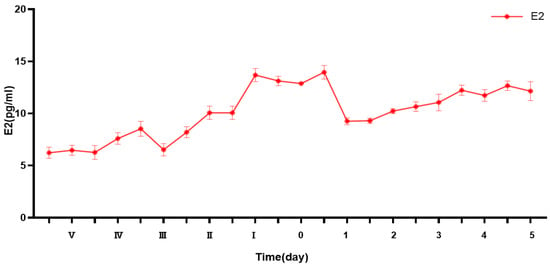
Figure 1
Open AccessArticle
AQP7-Mediated Mitochondrial Redox Homeostasis in Vitrified Oocytes: A Genetic Mechanism of PI3K/AKT Signaling Regulation
by
Yatian Qi, Wei Xia, Chenyu Tao, Xiaohuan Fang, Yang Yu, Jingwei Hu, Xiaofeng Tian, Tianmiao Qin, Congcong Yao, Wentao Zhang and Junjie Li
Genes 2025, 16(7), 730; https://doi.org/10.3390/genes16070730 - 23 Jun 2025
Abstract
Background/Objectives: Cellular oxidative stress is crucial for GV stage oocyte vitrification quality. PI3K and the aquaporin family have been shown to facilitate various cellular processes related to redox homeostasis and energy balance; yet, the mechanisms underlying the involvement of aquaporin 7 (AQP7) in
[...] Read more.
Background/Objectives: Cellular oxidative stress is crucial for GV stage oocyte vitrification quality. PI3K and the aquaporin family have been shown to facilitate various cellular processes related to redox homeostasis and energy balance; yet, the mechanisms underlying the involvement of aquaporin 7 (AQP7) in vitrified oocyte oxidative stress remain unclear. The purpose of the present investigation was to evaluate the role of AQP7 in vitrified oocytes and the mechanisms involved. Methods: AQP7 inhibitors were employed to investigate the effect of AQP7 on oxidative stress in mouse-vitrified oocytes, whereas PI3K activators were harnessed to ascertain whether AQP7 serves as a functional molecule involved in this process. Results: Our results indicate that AQP7 inhibition in vitrified oocytes results in a significant decrease in glutathione (GSH) levels associated with cellular oxidation and an elevation in H2O2 levels. This was accompanied by exacerbated mitochondrial dysfunction, weakened cytoskeletal proteins, accelerated early apoptosis. Consequently, both survival and maturation rates were markedly reduced. Interestingly, PI3K/AKT activation increased AQP7 expression, restored abnormal mitochondrial distribution, as well as calcium homeostasis, and rescued the oocyte survival/maturation rate. Conclusions: Our results provide new insights indicating that PI3K/AKT/AQP7 decreases oxidative stress by regulating mitochondrial morphology, function, and distribution, thereby rescuing oocyte maturation in vitrification.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
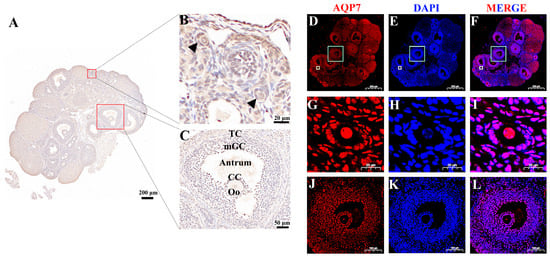
Figure 1
Open AccessArticle
Local Adaptation Shapes Phenotypic and Genetic Diversity in Zygophyllum loczyi
by
Jan-Cheng Wang, De-Yan Wu, Xue-Rong Li, Jia-Yi Lu, Suo-Min Wang, Qing Ma, Hai-Shuang Liu, Xi-Yong Wang, Jing-Dian Liu and Dao-Yuan Zhang
Genes 2025, 16(7), 729; https://doi.org/10.3390/genes16070729 - 23 Jun 2025
Abstract
Background/Objectives: Desert plants exhibit remarkable resilience to extreme environments, and their capacity for population establishment is noteworthy. However, the adaptation process mechanisms of those plants to harsh habitats, particularly concerning intraspecific differentiation and genetic diversity, remain poorly understood, and a comprehensive framework is lacking.
[...] Read more.
Background/Objectives: Desert plants exhibit remarkable resilience to extreme environments, and their capacity for population establishment is noteworthy. However, the adaptation process mechanisms of those plants to harsh habitats, particularly concerning intraspecific differentiation and genetic diversity, remain poorly understood, and a comprehensive framework is lacking. Zygophyllum loczyi Kanitz, an annual or biennial desert herb, demonstrates significant phenotypic plasticity across diverse habitats. Methods: Using mixed-effects models, this study examined 20 populations from four deserts to assess phenotypic variation and predict trait_environment relationships. Results: The findings indicated substantial inter-population phenotypic differentiation in Z. loczyi, with greater variation observed between deserts than within them. Traits such as blade length, petal length, sepal length, and stamen length were influenced by environmental conditions. Mixed-effects model prediction showed that the growth location of Z. loczyi significantly impacted its phenotypic traits. The characteristics of the four desert populations displayed varying responses to temperature and moisture changes, with the most pronounced response noted in the Gurbantunggut desert (Gt) population, indicating that survival stress has an important influence on the performance of plants. The single nucleotide polymorphisms result further confirmed that the differentiation and genetic diversity of the Gt population displayed the highest selection pressure, resulting the small effective size of the population. Conclusions: This study uncovers the adaptive mechanism of Z. loczyi to habitat through investigating the inter-population phenotypic differentiation and genetic diversity and provides new insight into local adaptation and evolutionary processes in the desert environment.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures
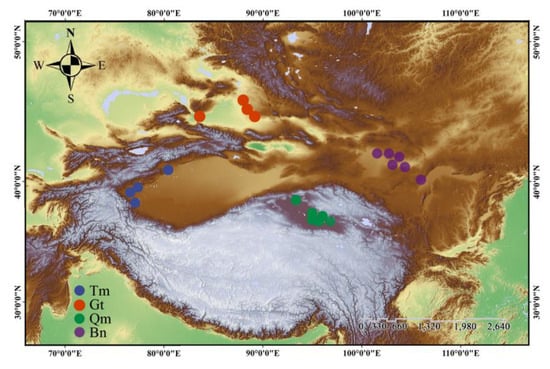
Figure 1

Journal Menu
► ▼ Journal Menu-
- Genes Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Diversity, Forests, Genes, IJPB, Plants
Plant Chloroplast Genome and Evolution
Topic Editors: Chao Shi, Lassaâd Belbahri, Shuo WangDeadline: 31 August 2025
Topic in
Biomolecules, IJMS, Marine Drugs, Molecules, Sci. Pharm., Genes, Pharmaceutics, Crystals
Bioinformatics in Drug Design and Discovery—2nd Edition
Topic Editors: Bing Niu, Suren Rao Sooranna, Pufeng DuDeadline: 30 September 2025
Topic in
Agriculture, Agronomy, Crops, Genes, Plants, DNA
Vegetable Breeding, Genetics and Genomics, 2nd Volume
Topic Editors: Umesh K. Reddy, Padma Nimmakayala, Georgia NtatsiDeadline: 31 October 2025
Topic in
Brain Sciences, CIMB, Epigenomes, Genes, IJMS, DNA
Genetics and Epigenetics of Substance Use Disorders
Topic Editors: Aleksandra Suchanecka, Anna Maria Grzywacz, Kszysztof ChmielowiecDeadline: 15 November 2025

Conferences
Special Issues
Special Issue in
Genes
Exploring Nucleolar Organization and Ribosomal Gene Mutations in Human Health and Disease
Guest Editor: Paola BellostaDeadline: 30 June 2025
Special Issue in
Genes
Genetics in Transplantation
Guest Editor: Brendan J. KeatingDeadline: 30 June 2025
Special Issue in
Genes
Genetics and Therapy of Retinal Diseases
Guest Editor: Yu Fujinami-YokokawaDeadline: 30 June 2025
Special Issue in
Genes
Genetic Diagnostics: Precision Tools for Disease Detection
Guest Editor: Honey ReddiDeadline: 30 June 2025
Topical Collections
Topical Collection in
Genes
Genotype-Phenotype Study in Disease
Collection Editors: Michele Cioffi, Maria Teresa Vietri
Topical Collection in
Genes
Feature Papers in ‘Animal Genetics and Genomics’
Collection Editors: Antonio Figueras, Raquel Vasconcelos








