Using STR Data to Investigate the Impact of the Studbook Cap on Genetic Diversity in the American Standardbred Horse from 1998 to 2021
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Analysis
3. Results
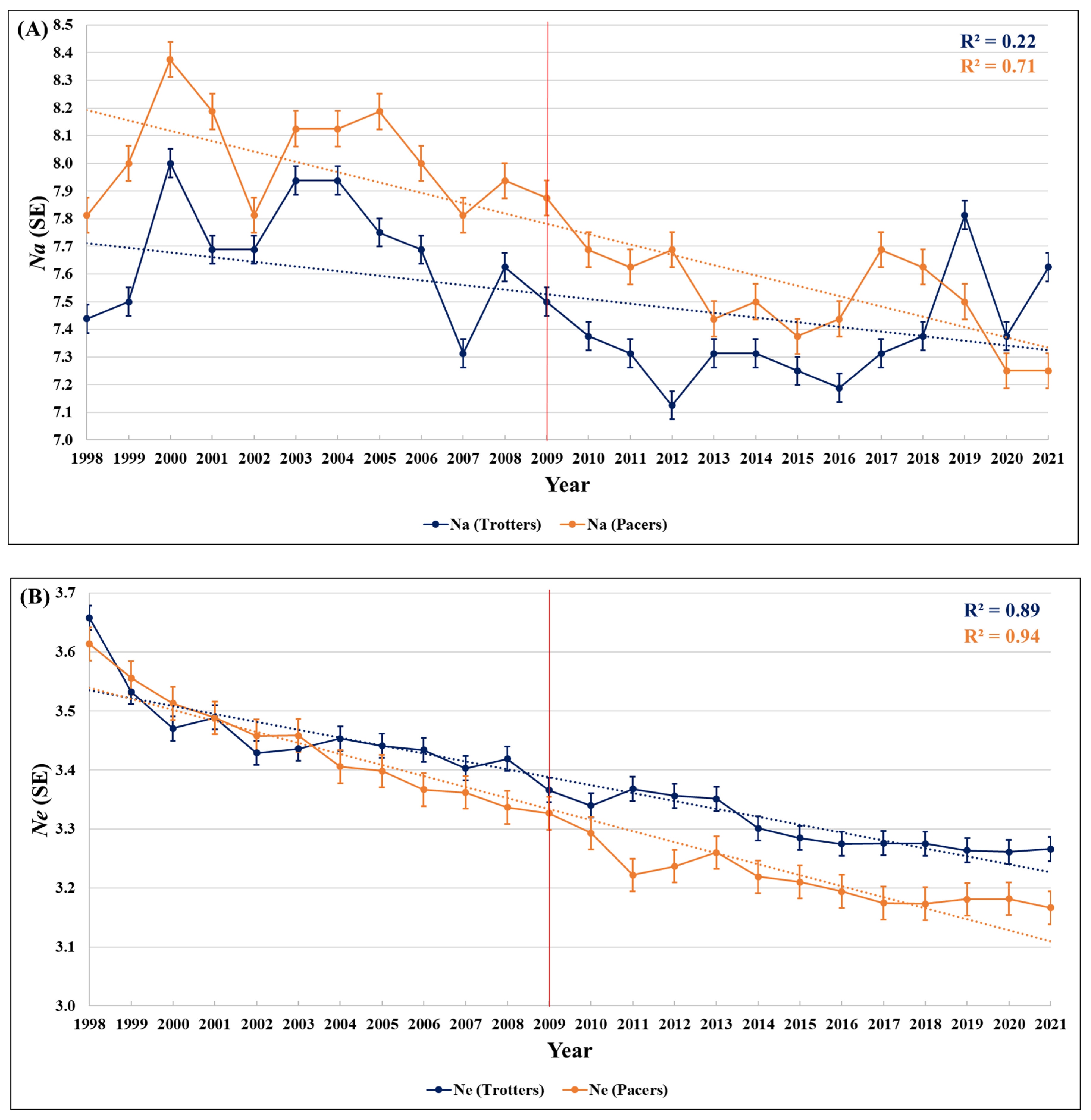
3.1. Allelic Richness (Na)
3.2. Effective Number of Alleles (Ne)
3.3. Expected Heterozygosity (HE)
3.4. Observed Heterozygosity (HO)
3.5. Inbreeding Coefficient (FIS)
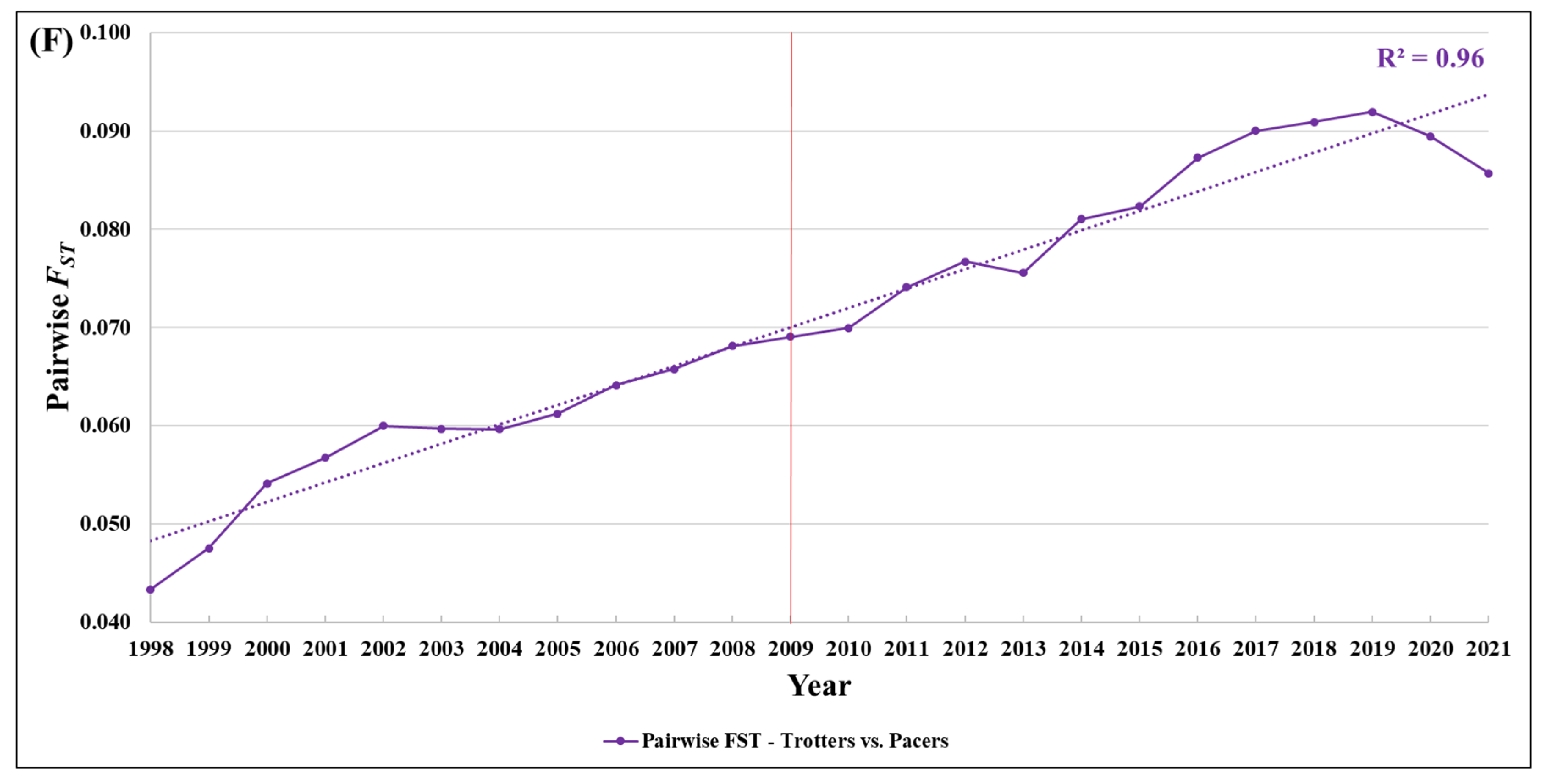
3.6. Pairwise Fixation Index (FST)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacCluer, J.W.; Boyce, A.J.; Dyke, B.; Weitkamp, L.R.; Pfenning, D.W.; Parsons, C.J. Inbreeding and pedigree structure in Standardbred horses. J. Hered. 1983, 74, 394–399. [Google Scholar] [CrossRef]
- Cothran, E.G.; MacCluer, J.W.; Weitkamp, L.R.; Guttormsen, S.A. Genetic variability, inbreeding, and reproductive performance in Standardbred horses. Zoo Biol. 1986, 5, 191–201. [Google Scholar] [CrossRef]
- Cothran, E.G.; MacCluer, J.W.; Weitkamp, L.R.; Pfennig, D.W.; Boyce, A.J. Inbreeding and reproductive performance in Standardbred horses. J. Hered. 1984, 75, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Cothran, E.G.; MacCluer, J.W.; Weitkamp, L.R.; Bailey, E. Genetic differentiation associated with gait within American Standardbred horses. Anim. Genet. 1987, 18, 285–296. [Google Scholar] [CrossRef] [PubMed]
- King, J.A. Changes in Heterozygosity Through Time in American Standardbred and American Saddlebred Horses (1960–1990). Master Thesis, University of North Texas, Denton, TX, USA, 1992. [Google Scholar]
- Petersen, J.L.; Mickelson, J.R.; Cothran, E.G.; Andersson, L.S.; Axelsson, J.; Bailey, E.; Bannasch, D.; Binns, M.M.; Borges, A.S.; Brama, P.; et al. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS ONE 2013, 8, e54997. [Google Scholar] [CrossRef] [PubMed]
- Esdaile, E.; Avila, F.; Bellone, R.R. Analysis of Genetic Diversity in the American Standardbred Horse Utilizing Short Tandem Repeats and Single Nucleotide Polymorphisms. J. Hered. 2022, 113, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Cho, G.J.; Cho, B.W. Microsatellite DNA Typing Using 16 Markers for Parentage Verification of the Korean Native Horse. Asian-Aust. J. Anim. Sci. 2004, 17, 750–754. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Cho, G.J. Standardization and Usefulness of ISAG Microsatellite Markers for Individual Identification and Parentage Verification in Horse Breeds. J. Vet. Clin. 2009, 26, 220–225. [Google Scholar]
- Peakall, R. & Smouse P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. Hierfstat, a package for R to compute and test variance components and F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 18 March 2024).
- Lenth, R. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R Package Version 1.8.4-1. 2023. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 18 March 2024).
- Wright, S. Evolution and the Genetics of Populations; University of Chicago Press: Chicago, IL, USA, 1978; Volume 4. [Google Scholar]
- Ellegren, H.; Johansson, M.; Sandberg, K.; Andersson, L. Cloning of highly polymorphic microsatellites in the horse. Anim. Genet. 1992, 23, 133–142. [Google Scholar] [CrossRef]
- Bowling, A.T.; Penedo, M.C.T.; Stott, M.L.; Malyj, W. Introduction of DNA Testing for Horse and Cattle Parentage Verification, Proceedings of the 5th International Symposium on Human Identification, Scottsdale, AZ, USA, 27–29 October 1993; Promega: Singapore, 1994; pp. 75–80. [Google Scholar]
- Binns, M.M.; Holmes, N.G.; Holliman, A.; Scott, A.M. The identification of polymorphic microsatellite loci in the horse and their use in Thoroughbred parentage testing. Br. Vet. J. 1995, 151, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Bowling, A.T.; Eggleston-Stott, M.L.; Byrns, G.; Clark, R.S.; Dileanis, S.; Wictum, E. Validation of microsatellite markers for routine horse parentage testing. Anim. Genet. 1997, 28, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Tozaki, T.; Kakoi, H.; Nakamura, K.; Rajabi-Toustani, R.; Ohba, Y.; Matsubara, T.; Takasu, M. Decreased genetic diversity in Kiso horses revealed through annual microsatellite genotyping. J. Vet. Med. Sci. 2020, 82, 503–505. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Oyungerel, B.; Kong, H.S. Genetic diversity and population structure of Mongolian regional horses with 14 microsatellite markers. Anim. Biosci. 2022, 35, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Orazymbetova, Z.; Ualiyeva, D.; Dossybayev, K.; Torekhanov, A.; Sydykov, D.; Mussayeva, A.; Baktybayev, G. Genetic diversity of Kazakhstani Equus caballus (Linnaeus, 1758) horse breeds inferred from microsatellite markers. Vet. Sci. 2023, 10, 598. [Google Scholar] [CrossRef]
- Yordanov, G.; Mehandjyiski, I.; Palova, N.; Atsenova, N.; Neov, B.; Radoslavov, G.; Hristov, P. Genetic diversity and structure of the main Danubian horse paternal genealogical lineages based on microsatellite genotyping. Vet. Sci. 2022, 9, 333. [Google Scholar] [CrossRef] [PubMed]
- Fornal, A.; Kowalska, K.; Zabek, T.; Piestrzynska-Kajtoch, A.; Musiał, A.D.; Ropka-Molik, K. Genetic diversity and population structure of Polish Konik horse based on individuals from all the male founder lines and microsatellite markers. Animals 2020, 10, 1569. [Google Scholar] [CrossRef] [PubMed]
- Khanshour, A.; Conant, E.; Juras, R.; Cothran, E.G. Microsatellite analysis of genetic diversity and population structure of Arabian horse populations. J. Hered. 2013, 104, 386–398. [Google Scholar] [CrossRef] [PubMed]

| Year | Trotters | Pacers |
|---|---|---|
| 1998 | 1145 | 2188 |
| 1999 | 2357 | 3882 |
| 2000 | 3785 | 5862 |
| 2001 | 3832 | 5866 |
| 2002 | 3835 | 5737 |
| 2003 | 3850 | 5907 |
| 2004 | 3986 | 5255 |
| 2005 | 3880 | 5300 |
| 2006 | 3915 | 4993 |
| 2007 | 3512 | 3640 |
| 2008 | 3540 | 4360 |
| 2009 | 3408 | 3908 |
| 2010 | 3344 | 4091 |
| 2011 | 3070 | 3830 |
| 2012 | 3100 | 3580 |
| 2013 | 2908 | 3470 |
| 2014 | 2971 | 3489 |
| 2015 | 3087 | 3211 |
| 2016 | 3033 | 3036 |
| 2017 | 3195 | 3014 |
| 2018 | 3512 | 2986 |
| 2019 | 3821 | 3049 |
| 2020 | 4073 | 3104 |
| 2021 | 3064 | 2443 |
| Total | 80,223 | 96,201 |
| Trotters | |||||
|---|---|---|---|---|---|
| Na | Ne | HO | HE | FIS | |
| Pre-studbook cap (1998–2009) mean | 7.67 | 3.46 | 0.68 | 0.68 | −0.01 |
| Post-studbook cap (2010–2021) mean | 7.36 | 3.30 | 0.67 | 0.66 | −0.01 |
| P-value | 1 × 10−3 | <1 × 10−5 | 1 × 10−4 | 1 × 10−4 | 1.3 × 10−1 |
| Pacers | |||||
| Na | Ne | HO | HE | FIS | |
| Pre-studbook cap (1998–2009) mean | 8.02 | 3.44 | 0.69 | 0.69 | 0.00 |
| Post-studbook cap (2010–2021) mean | 7.51 | 3.21 | 0.67 | 0.66 | −0.01 |
| P-value | <1 × 10−5 | <1 × 10−5 | <1 × 10−5 | <1 × 10−5 | 2.1 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila, F.; Esdaile, E.; Bellone, R.R. Using STR Data to Investigate the Impact of the Studbook Cap on Genetic Diversity in the American Standardbred Horse from 1998 to 2021. Genes 2025, 16, 748. https://doi.org/10.3390/genes16070748
Avila F, Esdaile E, Bellone RR. Using STR Data to Investigate the Impact of the Studbook Cap on Genetic Diversity in the American Standardbred Horse from 1998 to 2021. Genes. 2025; 16(7):748. https://doi.org/10.3390/genes16070748
Chicago/Turabian StyleAvila, Felipe, Elizabeth Esdaile, and Rebecca R. Bellone. 2025. "Using STR Data to Investigate the Impact of the Studbook Cap on Genetic Diversity in the American Standardbred Horse from 1998 to 2021" Genes 16, no. 7: 748. https://doi.org/10.3390/genes16070748
APA StyleAvila, F., Esdaile, E., & Bellone, R. R. (2025). Using STR Data to Investigate the Impact of the Studbook Cap on Genetic Diversity in the American Standardbred Horse from 1998 to 2021. Genes, 16(7), 748. https://doi.org/10.3390/genes16070748






