-
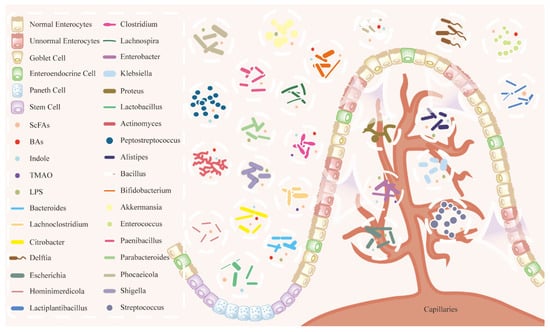
 Neutrophils at the Crossroads of Oral Microbiome Dysbiosis and Periodontal Disease
Neutrophils at the Crossroads of Oral Microbiome Dysbiosis and Periodontal Disease -
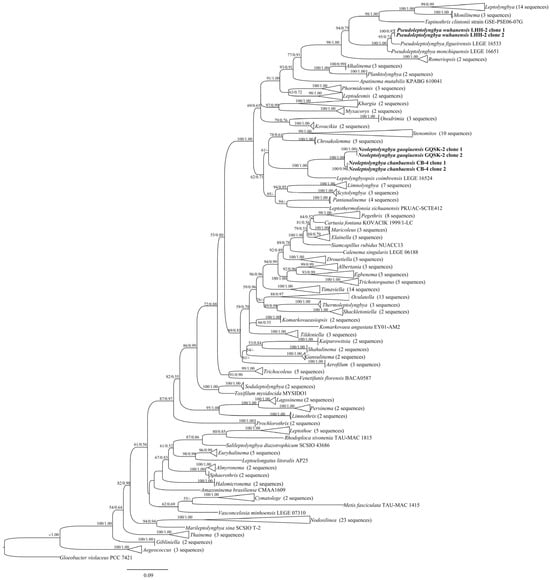
 Interaction of Bacteria and Fleas, Focusing on the Plague Bacterium—A Review
Interaction of Bacteria and Fleas, Focusing on the Plague Bacterium—A Review -
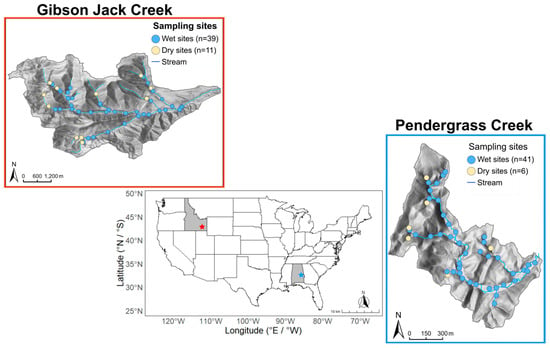
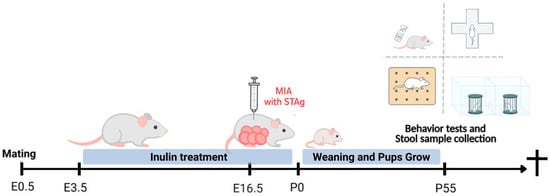
 Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts
Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts -
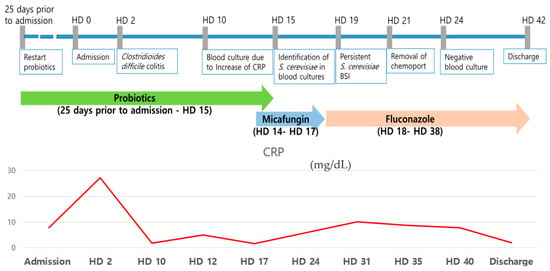
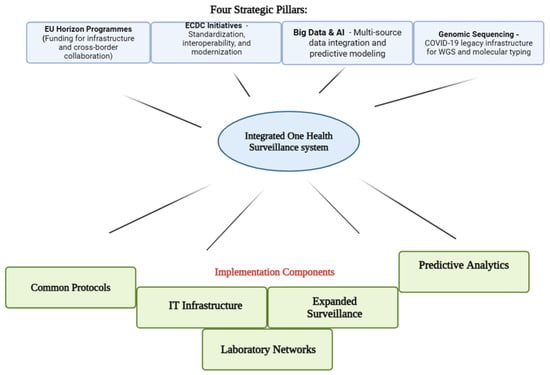
 Distinct Seasonal Patterns: Three Years (2022–2025) of Wastewater-Based Influenza Surveillance in Munich
Distinct Seasonal Patterns: Three Years (2022–2025) of Wastewater-Based Influenza Surveillance in Munich -
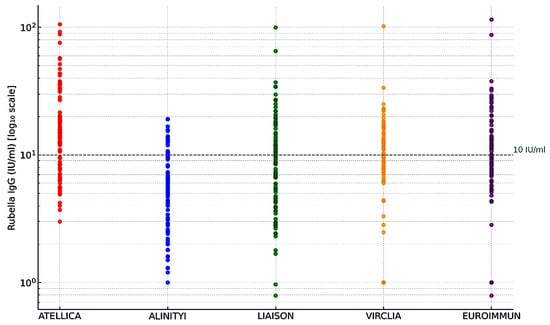
 Antimicrobial Peptides: Current Status, Mechanisms of Action, and Strategies to Overcome Therapeutic Limitations
Antimicrobial Peptides: Current Status, Mechanisms of Action, and Strategies to Overcome Therapeutic Limitations
Journal Description
Microorganisms
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, PubAg, CAPlus / SciFinder, AGRIS, and other databases.
- Journal Rank: JCR - Q2 (Microbiology) / CiteScore - Q1 (Microbiology (medical))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 15.2 days after submission; acceptance to publication is undertaken in 2.9 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Testimonials: See what our editors and authors say about Microorganisms.
- Companion journal for Microorganisms include: Applied Microbiology and Bacteria.
Latest Articles
E-Mail Alert
News
Topics
Deadline: 31 December 2025
Deadline: 31 January 2026
Deadline: 1 February 2026
Deadline: 28 February 2026
Conferences
Special Issues
Deadline: 31 December 2025
Deadline: 31 December 2025
Deadline: 31 December 2025
Deadline: 31 December 2025