-
 Identification and Characterization of MmuPV1 Causing Papillomatosis Outbreak in an Animal Research Facility
Identification and Characterization of MmuPV1 Causing Papillomatosis Outbreak in an Animal Research Facility -
 Potential for Core Fucose-Targeted Therapy Against HBV Infection of Human Normal Hepatocytes
Potential for Core Fucose-Targeted Therapy Against HBV Infection of Human Normal Hepatocytes -
 Broadly Sarbecovirus-Neutralizing Antibodies Induced by Ancestral SARS-CoV-2 Infection
Broadly Sarbecovirus-Neutralizing Antibodies Induced by Ancestral SARS-CoV-2 Infection -
 Dysregulation of microRNAs in the Brains of Mice Infected with Powassan Virus
Dysregulation of microRNAs in the Brains of Mice Infected with Powassan Virus -
 Immune Responses and Replication of Rescued Torque Teno Virus (TTSuV1) in Mice
Immune Responses and Replication of Rescued Torque Teno Virus (TTSuV1) in Mice
Journal Description
Viruses
Viruses
is a peer-reviewed, open access journal of virology, published monthly online by MDPI. The Spanish Society for Virology (SEV), Canadian Society for Virology (CSV), Italian Society for Virology (SIV-ISV), Australasian Virology Society (AVS), Brazilian Society for Virology (BSV) and others are affiliated with Viruses and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Virology) / CiteScore - Q1 (Virology/Infectious Diseases)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 18.6 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Companion journal: Zoonotic Diseases.
Impact Factor:
3.5 (2024);
5-Year Impact Factor:
3.7 (2024)
Latest Articles
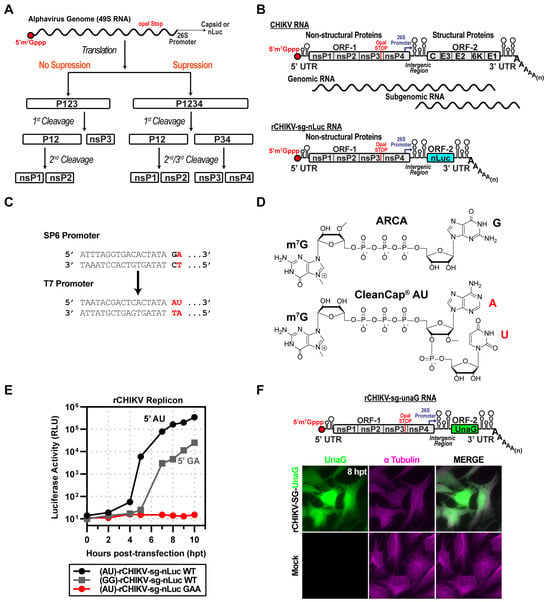
Minimal Polymerase-Containing Precursor Required for Chikungunya Virus RNA Synthesis
Viruses 2025, 17(12), 1556; https://doi.org/10.3390/v17121556 (registering DOI) - 28 Nov 2025
Abstract
Alphaviruses pose a growing global health threat, with Chikungunya virus (CHIKV) epidemics ongoing. Although several CHIKV vaccine candidates have progressed to late-stage clinical evaluation, none have yet achieved licensure or widespread availability. The CHIKV nonstructural proteins nsP2 and nsP4 encode essential enzymatic activities
[...] Read more.
Alphaviruses pose a growing global health threat, with Chikungunya virus (CHIKV) epidemics ongoing. Although several CHIKV vaccine candidates have progressed to late-stage clinical evaluation, none have yet achieved licensure or widespread availability. The CHIKV nonstructural proteins nsP2 and nsP4 encode essential enzymatic activities that represent key targets for antiviral development, yet the biochemical basis of nsP4 RNA-dependent RNA polymerase (RdRp) activity remains poorly understood. Here, we identify a minimal, functional precursor form of nsP4 derived from the nsP3–nsP4 polyprotein (P34) that is active in a cell-based RNA replicon system. Using synthetic, capped mRNAs, we show that cleavage of P34 by the nsP2 protease is required for robust reporter expression, and that a truncated form retaining only the C-terminal 50 residues of nsP3 (CT50-P34) supports near-wild-type replication. Unexpectedly, ubiquitin–nsP4 fusions failed to substitute for P34, likely reflecting the transient expression supported by our RNA-based system. We propose that precursor forms of nsP4 interact with the nsP1 dodecamer at the site of genome replication, where cleavage activates the RdRp and localization within the nsP1 dodecamer maintains nsP4 in its active conformation. Dissociation from the nsP1 dodecamer triggers a conformational switch to an inactive state. Together, these findings establish a tractable framework for interrogation of the assembly, activation, and regulation of the alphavirus polymerase.
Full article
(This article belongs to the Special Issue 15-Year Anniversary of Viruses)
►
Show Figures
Open AccessReview
Virus Diseases of Economic Importance on Food Legumes in Africa and Their Control
by
Adane Abraham
Viruses 2025, 17(12), 1555; https://doi.org/10.3390/v17121555 - 28 Nov 2025
Abstract
Virus diseases are among the major constraints in the production of food legumes in Africa, causing substantial crop losses. Common bean mosaic and black root, cowpea mosaic, chickpea stunt, faba bean necrotic yellows and stunt, groundnut rosette, and soybean mosaic are the six
[...] Read more.
Virus diseases are among the major constraints in the production of food legumes in Africa, causing substantial crop losses. Common bean mosaic and black root, cowpea mosaic, chickpea stunt, faba bean necrotic yellows and stunt, groundnut rosette, and soybean mosaic are the six diseases considered economically significant in Africa. Past research enabled the description of the main characteristics of the causal viruses, including particle and genome properties, modes of transmission, host range, and virus–vector relationships. Such information in many cases assisted in developing effective diagnostics and disease management methods such as host resistance, chemical vector control, and cultural practices. Integrating two or more of these approaches is usually more effective. The major challenge, however, remains ensuring the adoption of such recommendations at a sufficiently large scale by many farmers to have an impact over wider geographical areas. Future work should focus on scaling up the adoption of available control technologies and generating new information, including epidemiological data, to support future management decisions. Furthermore, since the occurrence and significance of viruses on food legumes in many African countries are still not studied, large-scale surveys to identify viruses, establish their distribution and impact, and working out suitable control measures are required.
Full article
(This article belongs to the Special Issue Economically Important Viruses in African Crops)
►▼
Show Figures
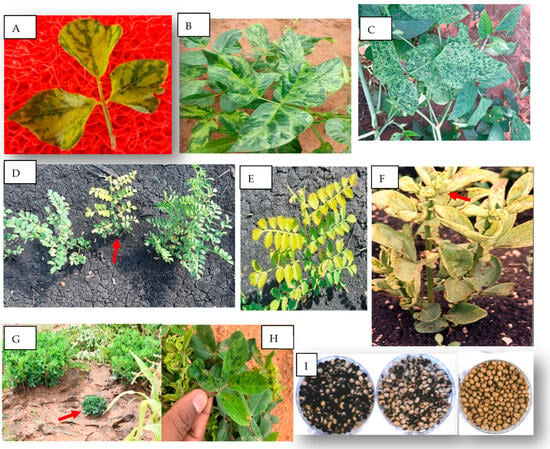
Figure 1
Open AccessReview
Acute Respiratory Infections (ARIs): Current Etiological Perspectives and Advances in Viral Metagenomics—A Review
by
Murilo Marconi dos Santos, Tiago Souza Salles, Gabriel Montenegro de Campos, Isabela Carvalho Brcko, Alex Ranieri Jerônimo Lima, Sandra Coccuzzo Sampaio, Maria Carolina Elias, Marta Giovanetti and Svetoslav Nanev Slavov
Viruses 2025, 17(12), 1554; https://doi.org/10.3390/v17121554 - 27 Nov 2025
Abstract
Acute respiratory infections (ARIs) remain a leading cause of global morbidity and mortality, disproportionately affecting vulnerable populations such as children, the elderly, and immunocompromised individuals. Despite the availability of traditional diagnostic tools, including viral culture and highly elaborated PCR respiratory panels, many cases
[...] Read more.
Acute respiratory infections (ARIs) remain a leading cause of global morbidity and mortality, disproportionately affecting vulnerable populations such as children, the elderly, and immunocompromised individuals. Despite the availability of traditional diagnostic tools, including viral culture and highly elaborated PCR respiratory panels, many cases of ARI remain without an identified etiological agent. This is due to the vast diversity of viral agents that can be involved in cases of ARI, which represents a major limitation of the targeted diagnosis. In this context, viral metagenomics has emerged as a powerful, unbiased approach for detecting both known and novel pathogens directly from clinical samples. This review highlights the application of metagenomic next-generation sequencing for the investigation of etiological causes of ARIs, emphasizing its relevance in complex cases—particularly among immunocompromised patients—where standard methods might fail. We highlight the main viruses involved in respiratory infections, the strengths and limitations of metagenomic next-generation sequencing approaches, their role in genomic surveillance of respiratory viruses, and their potential to build public health responses to potentially emerging respiratory threats. Ultimately, integrating viral metagenomics into clinical and surveillance frameworks could enhance the early detection and control of respiratory viral diseases worldwide.
Full article
(This article belongs to the Special Issue Influenza and Other Respiratory Viruses: Prevention, Diagnosis, Treatment: 2nd Edition)
►▼
Show Figures
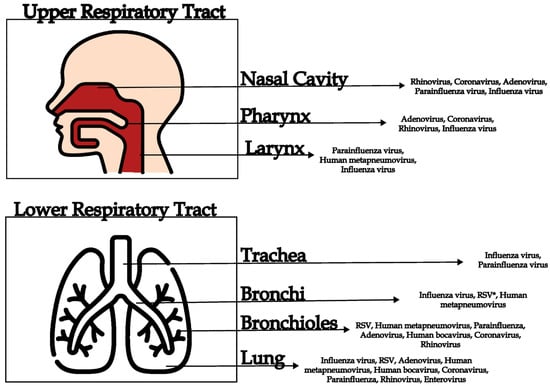
Figure 1
Open AccessArticle
A Feasibility Study of a Rapid Lateral Flow Immunoassay Test for Children and Adolescents to Determine SARS-CoV-2 Seroprevalence
by
Rukiye Saç, Medine Ayşin Taşar, Burcu Ceylan Cura Yayla, Serçin Taşar, Meliha Sevim, Gözde Onay and İlknur Fidancı
Viruses 2025, 17(12), 1553; https://doi.org/10.3390/v17121553 - 27 Nov 2025
Abstract
Global seroprevalence of SARS-CoV-2 in children and adolescents has risen to an estimated 56.6%. Limited data on children highlights the need for ongoing surveillance as recommended by the WHO. Understanding this need, in this study we carry out a feasibility study of a
[...] Read more.
Global seroprevalence of SARS-CoV-2 in children and adolescents has risen to an estimated 56.6%. Limited data on children highlights the need for ongoing surveillance as recommended by the WHO. Understanding this need, in this study we carry out a feasibility study of a rapid lateral flow immunoassay test (LFIAT) and determine SARS-CoV-2 seroprevalence in children and adolescents. The method used in the paper included 202 children (1–18 years old) who were evaluated for SARS-CoV-2 IgG/IgM antibodies using the LFIAT. LFIAT provides results in 20 min. Demographic data was collected through questionnaires. As a result, SARS-CoV-2 antibody seroprevalence was 52.0% (n = 105). No gender differences were observed. Adolescents exhibited higher seroprevalence (63.0%) compared to children aged 1–5.9 (49.3%) and 6–11.9 years old (46.8%), without statistical significance (p > 0.05). The median age of seropositive children was 9 years, and 77.1% had a history of PCR-confirmed infection (p = 0.001). In conclusion, results are consistent with global data. It was observed that the LFIAT can be easily administered to children and parents had a positive outlook on the test. Findings suggest that LFIAT can be used to assess SARS-CoV-2 immunity, while providing the first seroprevalence estimate for SARS-CoV-2 among healthy children and adolescents in Turkey, utilizing the COVID-19 IgG/IgM Rapid Test-KitA.
Full article
(This article belongs to the Section Coronaviruses)
Open AccessArticle
Comparative Assessment of Viral Load Retention in Surgical and Fabric Masks Worn by COVID-19 Patients
by
Cristiane Monteiro Eller, Milena De Paula Rebello, Andreza Sálvio, Emanuelle S. R. F. Silva, Vinícius Silva Belo, Elba Regina E. Lemos, Marta Giovanetti, José Júnior França De Barros and Marco Aurélio Horta
Viruses 2025, 17(12), 1552; https://doi.org/10.3390/v17121552 - 27 Nov 2025
Abstract
Face masks are widely recognized as a key intervention to limit SARS-CoV-2 transmission, yet the distribution and persistence of viral RNA across different mask regions and layers remain poorly understood. To address this, we analyzed 185 masks collected from 60 SARS-CoV-2-positive individuals in
[...] Read more.
Face masks are widely recognized as a key intervention to limit SARS-CoV-2 transmission, yet the distribution and persistence of viral RNA across different mask regions and layers remain poorly understood. To address this, we analyzed 185 masks collected from 60 SARS-CoV-2-positive individuals in Rio de Janeiro between December 2020 and September 2022. Masks were sectioned into anatomical regions (nose, mouth, sides) and structural layers (inner, middle, outer), and viral RNA was quantified using RT-qPCR. Samples with the highest viral loads were selected for partial sequencing of the spike gene, and paired analyses with swab samples were performed. Statistical comparisons included non-parametric tests and a linear mixed-effects model. Our results showed that the inner layer and nose region consistently harbored the highest viral RNA levels, with no significant differences between surgical and fabric masks. Viral load decreased by an estimated 39% per day, consistent with exponential decay. Sequencing confirmed identical viral genomes in masks and swabs and allowed identification of circulating variants, including Gamma and Omicron. These findings indicate that masks serve not only as effective physical barriers but also as non-invasive sources for genomic surveillance, providing insights into viral shedding patterns and informing strategies for monitoring and controlling SARS-CoV-2 transmission.
Full article
(This article belongs to the Special Issue Molecular Epidemiology of SARS-CoV-2, 4th Edition)
►▼
Show Figures

Graphical abstract
Open AccessArticle
Fatigue Severity, Cognitive Strain, and Psychological Health in Long COVID: Untangling the Interconnected Aftermath from a Dedicated Long COVID Clinic
by
Ashish Bhargava, Hemang Patel, Susan Szpunar, Mamta Sharma, Michael Somero, Shyam Moudgil and Louis Saravolatz
Viruses 2025, 17(12), 1551; https://doi.org/10.3390/v17121551 - 27 Nov 2025
Abstract
Post-acute sequelae of SARS-CoV-2 infection (PASC) frequently includes persistent fatigue and cognitive dysfunction, but the relationship between these symptoms remains poorly defined. In this prospective observational study at the Henry Ford St. John Long COVID Clinic (LCC) from July 2023 to March 2025,
[...] Read more.
Post-acute sequelae of SARS-CoV-2 infection (PASC) frequently includes persistent fatigue and cognitive dysfunction, but the relationship between these symptoms remains poorly defined. In this prospective observational study at the Henry Ford St. John Long COVID Clinic (LCC) from July 2023 to March 2025, we assessed fatigue severity using the Fatigue Assessment Scale (FAS) and examined its relationship with depression and cognitive symptoms. New patients completed demographic and clinical questionnaires, Patient Health Questionnaire (PHQ)-9, and Montreal Cognitive Assessment (MoCA) at their first LCC visit. Among 41 patients, 35 (85.4%) met the inclusion criteria for fatigue (FAS ≥ 22), with 18 (51.5%) experiencing severe fatigue (FAS > 34). Severe fatigue was significantly associated with shortness of breath, chest pain, and depression. Patients experiencing severe fatigue had significantly higher median PHQ-9 scores (12.5) compared to those with mild to moderate fatigue (5.0, p < 0.001). However, there were no significant differences in MoCA scores between these groups. Our study suggests a strong relationship between fatigue and depression in patients with PASC, emphasizing the importance of integrated physical and psychological healthcare. Moreover, since cognitive performance does not vary with fatigue levels, all PASC patients with cognitive dysfunction should receive routine cognitive screenings, regardless of the severity of their fatigue.
Full article
(This article belongs to the Section Coronaviruses)
Open AccessArticle
Molecular Determinants of Species-Specific Interactions Between Protein Kinase R and Poxvirus K3 Orthologs
by
Chorong Park, Greg Brennan, Chen Peng, Chi Zhang, Jingxin Cao, Loubna Tazi and Stefan Rothenburg
Viruses 2025, 17(12), 1550; https://doi.org/10.3390/v17121550 - 26 Nov 2025
Abstract
Protein kinase R (PKR) is an antiviral protein that is involved in molecular “arms races” with viral antagonists. As a result, some PKR inhibitors, including the vaccinia virus (VACV) protein K3 and its orthologs from other poxviruses only inhibit PKRs of selected species.
[...] Read more.
Protein kinase R (PKR) is an antiviral protein that is involved in molecular “arms races” with viral antagonists. As a result, some PKR inhibitors, including the vaccinia virus (VACV) protein K3 and its orthologs from other poxviruses only inhibit PKRs of selected species. We previously reported contrasting inhibition patterns of human, sheep, and cow PKRs by VACV K3 and the sheeppox virus (SPPV) K3 ortholog, SPPV 011. Here we show that the differential sensitivities of cow and sheep PKRs to VACV K3 were mediated by only two residues in PKR helix αG. In contrast, SPPV 011 sensitivities were governed by additional residues and regions. Analysis of the PKR sensitivities from 20 mammalian species to VACV K3 and SPPV 011 revealed four different sensitivity patterns: some PKRs were inhibited by only one K3 ortholog, as previously reported, whereas other PKRs were either resistant or sensitive to both inhibitors. Furthermore, we characterized a residue (K45) in VACV K3 that is involved in the species-specific inhibition of PKR. Mutating this residue increased the inhibition of sheep but not human PKR, whereas it decreased the inhibition of mouse PKR, highlighting that a single mutation in a viral protein can result in distinct species-dependent inhibition changes.
Full article
(This article belongs to the Special Issue Viral Strategies and Cellular Countermeasures That Regulate mRNA Access to the Translation Apparatus: 2nd Edition)
►▼
Show Figures
Graphical abstract
Open AccessArticle
Molecular Aspects of the Emergence of Monkeypox Virus Clades
by
Igor V. Babkin, Irina N. Babkina and Nina V. Tikunova
Viruses 2025, 17(12), 1549; https://doi.org/10.3390/v17121549 - 26 Nov 2025
Abstract
Monkeypox virus (MPXV), which previously caused mainly zoonotic infections, is currently the causative agent of the mpox outbreak that began in 2022. Since the mpox outbreak is characterized by sustained human-to-human transmission, the evolutionary trajectory of MPXV is an important scientific issue. The
[...] Read more.
Monkeypox virus (MPXV), which previously caused mainly zoonotic infections, is currently the causative agent of the mpox outbreak that began in 2022. Since the mpox outbreak is characterized by sustained human-to-human transmission, the evolutionary trajectory of MPXV is an important scientific issue. The prevailing hypothesis suggests that the modern orthopoxviruses originated from cowpox-like ancestors with larger genomes that infected a wide range of hosts. Subsequent evolution included the reduction of the genome and the accumulation of substitutions in key proteins. Molecular dating of MPXV evolution revealed 5–6-fold acceleration in the evolutionary rate that was observed in subclade IIb after 2018, reaching 1.8 × 10−5 substitutions/site/year, likely due to virus’ adaptation to humans. The origin of MPXV from its precursor was primarily driven by the accumulation of non-synonymous substitutions in the key host range genes, including those associated with the protein inhibiting host protein synthesis (OPG173) and host immune evasion (OPG027). The subsequent divergence of MPXV into clades I and II largely depended on mutations in the gene encoding the Bcl-2-like protein. Finally, the division of clade II into subclades IIa and IIb was facilitated by further non-synonymous substitutions in the soluble interferon alpha/beta receptor and hemagglutinin genes.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures

Figure 1
Open AccessReview
Transmission, Pathological and Clinical Manifestations of Highly Pathogenic Avian Influenza A Virus in Mammals with Emphasis on H5N1 Clade 2.3.4.4b
by
Sandra Vibeke Larsen, Rebekka Israelson, Charlotte Torp, Lars Erik Larsen, Henrik Elvang Jensen and Charlotte Kristensen
Viruses 2025, 17(12), 1548; https://doi.org/10.3390/v17121548 - 26 Nov 2025
Abstract
Highly pathogenic avian influenza A virus (HPAIV) H5N1, clade 2.3.4.4b, has emerged as a significant zoonotic threat. H5N1 is widely circulating in wild birds, and an increasing number of spillover events have been observed in a wide range of mammalian species. These cases
[...] Read more.
Highly pathogenic avian influenza A virus (HPAIV) H5N1, clade 2.3.4.4b, has emerged as a significant zoonotic threat. H5N1 is widely circulating in wild birds, and an increasing number of spillover events have been observed in a wide range of mammalian species. These cases are primarily reported in countries on the European and American continents. This review describes the likely transmission routes, lesions, and clinical manifestations of HPAIV H5N1 clade 2.3.4.4b in naturally infected mammals, with a focus on the involvement of the central nervous system (CNS). In the analysis, pathological findings were categorized by organ system and host species, which were further divided into terrestrial mammals, marine mammals, and dairy cattle. The most frequently reported clinical manifestations were neurological and respiratory signs in marine mammals and neurological signs and lethargy in terrestrial mammals. Macroscopic and histological lesions were commonly found in the CNS and lungs of terrestrial and marine mammals, while dairy cattle showed mainly gastrointestinal and mammary gland involvement. Immunohistochemistry and reverse transcriptase real-time PCR analyses confirmed high viral loads in brain tissues, indicating a neurological tropism of H5N1 clade 2.3.4.4b. Routes of CNS invasion remain uncertain, though both hematogenous and olfactory nerve pathways are discussed. Recent evidence suggests mammal-to-mammal and vertical transmission, raising concerns for the zoonotic and pandemic potential of this virus. In conclusion, the findings emphasize an urgent need for enhanced surveillance to effectively disclose changes in viral pathogenicity and transmissibility among mammalian hosts.
Full article
(This article belongs to the Special Issue Molecular Epidemiology, Evolution, and Transmission of Avian Influenza Viruses)
►▼
Show Figures
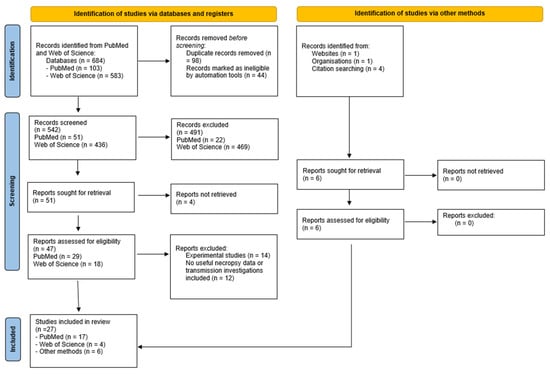
Figure 1
Open AccessArticle
Molecular Survey and Genetic Characterization of Hop Stunt Viroid (HSVd) in Fruit Trees in Kazakhstan
by
Leila T. Nadirova, Gulshan E. Stanbekova, Anna S. Nizkorodova, Ruslan V. Kryldakov, Bulat K. Iskakov and Andrey V. Zhigailov
Viruses 2025, 17(12), 1547; https://doi.org/10.3390/v17121547 - 26 Nov 2025
Abstract
Hop stunt viroid (HSVd) infects a variety of natural hosts, including fruit trees, leading to significant economic losses worldwide. This survey aimed to assess the incidence of HSVd in fruit trees in southern Kazakhstan. Out of 482 fruit trees examined, 28 (5.81%) were
[...] Read more.
Hop stunt viroid (HSVd) infects a variety of natural hosts, including fruit trees, leading to significant economic losses worldwide. This survey aimed to assess the incidence of HSVd in fruit trees in southern Kazakhstan. Out of 482 fruit trees examined, 28 (5.81%) were found to be infected with HSVd. The incidence was significantly higher in stone fruit trees (8.15%; 22/270) compared to pome fruit trees (2.82%; 6/212; p = 0.0133). Apricots had the highest infection rate at 12.66% (10/79), while pears had the lowest rate at 2.08% (1/48). Fifteen of the identified viroids were cloned for full-genome sequencing. Sequence analysis revealed a high percentage of nucleotide sequence similarity (99–100%) among the Kazakhstani HSVd isolates, suggesting a possible unique origin for the infection. We also identified several SNPs of HSVd that have not been previously documented. A phylogenetic analysis indicated that the Kazakhstani HSVd variants clustered together in a separate group, distinct from the five known groups of HSVd. This potentially new group displayed differences in its predicted secondary structure compared to other viroid groups. These findings emphasize the need for the development of effective control measures for HSVd and other viroids affecting fruit trees.
Full article
(This article belongs to the Section Viruses of Plants, Fungi and Protozoa)
►▼
Show Figures
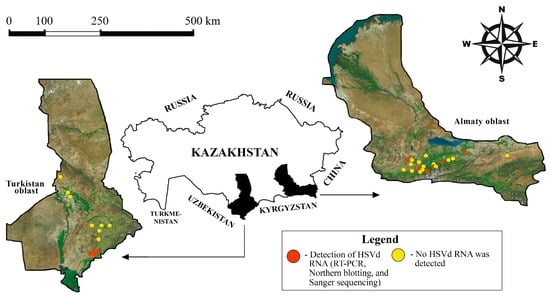
Figure 1
Open AccessArticle
Molecular and Phylogenetic Analyses of Lumpy Skin Disease Virus (LSDV) Outbreak (2021/22) in Pakistan Indicate Involvement of a Clade 1.2 LSDV Strain
by
Saiba Ferdoos, Andy Haegeman, Sadia Sattar, Ibrar Ahmed, Sundus Javed, Aamira Tariq, Nick De Regge and Nazish Bostan
Viruses 2025, 17(12), 1546; https://doi.org/10.3390/v17121546 - 26 Nov 2025
Abstract
Livestock is the backbone of the economy in an agricultural country like Pakistan, with cattle serving as a milk and protein source. In 2021/22, Pakistan was hit by the first major outbreak of lumpy skin disease (LSD) in cattle, in all four provinces.
[...] Read more.
Livestock is the backbone of the economy in an agricultural country like Pakistan, with cattle serving as a milk and protein source. In 2021/22, Pakistan was hit by the first major outbreak of lumpy skin disease (LSD) in cattle, in all four provinces. LSD is characterized by the development of skin nodules, leading to severe illness, decreased milk production, and mortality, causing huge economic losses. This study aimed to analyze and classify the lumpy skin disease virus (LSDV) strains involved in the outbreak in the Punjab province at the molecular and phylogenetic levels to develop effective control strategies. A combination of different real-time PCRs was used for the identification and differentiation between vaccine, wild-type, and recombinant LSDV strains. This was mented with the sequence determination and phylogenetic analysis of ten genomic loci from two selected isolates from the 2021/22 Pakistan outbreak. The combined data showed that these isolates belonged to LSDV clade 1.2 and were clearly different from the vaccine clade 1.1 (Neethling-like), as well as from the recombinant clade 2 strains. In addition, using a fit-for-purpose gel-based PCR, the isolates from the outbreak were also shown to be different from KSGP0240-based vaccines.
Full article
(This article belongs to the Special Issue Epidemiology, Risk Analysis, Detection and Management of Lumpy Skin Disease in Europe)
►▼
Show Figures
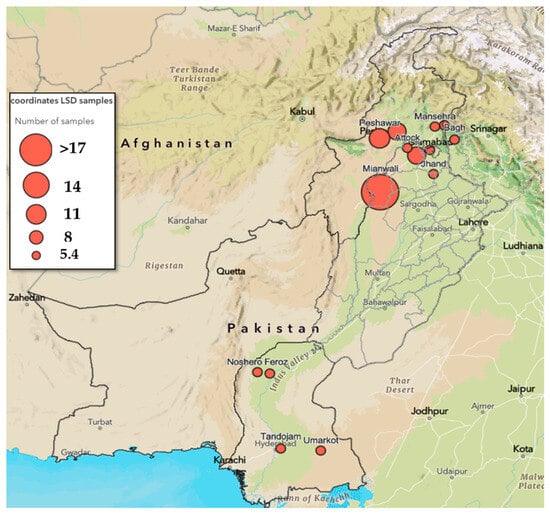
Figure 1
Open AccessArticle
Comparative Clinical Outcomes of Major Respiratory Viruses in Hospitalized Adults During the Post-Pandemic Period: A Retrospective Cohort Study
by
Hasip Kahraman, Gizem Keser, Furkan Süha Ölmezoğlu, Betül Altıntaş Öner, Onur Sedat Kurt, Tercan Us and Fatma Erdem
Viruses 2025, 17(12), 1545; https://doi.org/10.3390/v17121545 - 26 Nov 2025
Abstract
Background: In the post-pandemic era, respiratory viruses continue to cause substantial morbidity and mortality among hospitalized adults. SARS-CoV-2 and influenza remain the most common pathogens, while RSV and rhinovirus have re-emerged as relevant causes of severe illness. This study compared the characteristics and
[...] Read more.
Background: In the post-pandemic era, respiratory viruses continue to cause substantial morbidity and mortality among hospitalized adults. SARS-CoV-2 and influenza remain the most common pathogens, while RSV and rhinovirus have re-emerged as relevant causes of severe illness. This study compared the characteristics and outcomes of virus-specific infections detected by multiplex real-time PCR over two consecutive seasons. Methods: This retrospective cohort study was conducted at a 1010-bed tertiary-care hospital in Türkiye between June 2022 and June 2024. Adults hospitalized with at least one respiratory virus detected by MRT-PCR were included. Demographic, clinical, and laboratory data were analyzed. Pathogen-specific comparisons were limited to monoinfections, and predictors of in-hospital mortality were identified using multivariable logistic regression. Results: Among 518 admissions, influenza (33.6%) and SARS-CoV-2 (29.3%) were the predominant pathogens, followed by rhinovirus (11.2%), RSV (6.6%), and other respiratory viruses (19.6%). Overall in-hospital mortality was 26.6%. Mortality differed across virus groups in unadjusted analyses, being highest in SARS-CoV-2 and RSV and lowest in rhinovirus. Non-survivors were older, more comorbid, more often immunosuppressed, and more likely to require oxygen therapy or ICU care at sampling. In multivariable analysis, independent predictors of mortality were ICU location at sampling (aOR 5.52), oxygen requirement (aOR 3.39), immunosuppression (aOR 3.67), older age (per 10-year increase: aOR 1.25), and secondary bacterial infection (aOR 7.00). Viral etiology, including SARS-CoV-2, was not independently associated with mortality after adjustment. Conclusions: Among hospitalized adults, mortality was driven primarily by host-related factors and secondary bacterial infection rather than by viral etiology. These findings highlight the need for strengthened adult immunization programs, reliable respiratory virus surveillance, the prevention of bacterial superinfection, and the development of and equitable access to effective vaccines and antiviral therapies to reduce severe outcomes in high-risk adults.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures
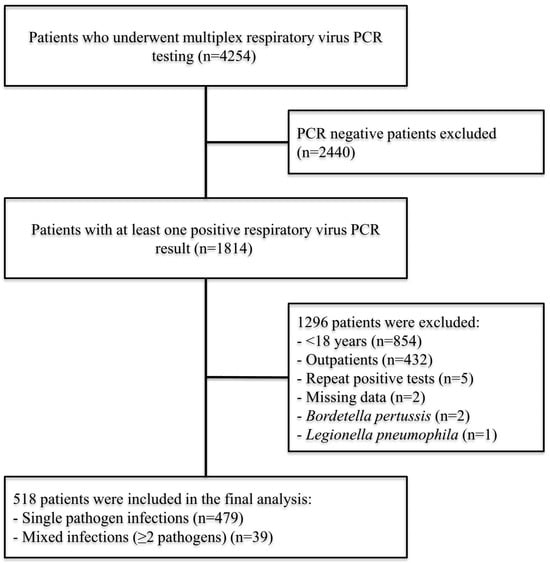
Figure 1
Open AccessArticle
Detection of Lymphocytic Choriomeningitis Virus in House Mouse (Mus musculus) in Brazil
by
Gabriel Rosa Cavalcanti, Jorlan Fernandes, Fernando de Oliveira Santos, Bernardo Rodrigues Teixeira, Alexandro Guterres, Julia Brignone, Silvana Levis, Camila dos Santos Lucio, Sócrates Fraga da Costa-Neto, Vagner Fonseca, Marta Giovanetti, Luiz Carlos Junior Alcantara, Paulo Sérgio D’Andrea, Elba Regina Sampaio de Lemos and Renata Carvalho de Oliveira
Viruses 2025, 17(12), 1544; https://doi.org/10.3390/v17121544 - 26 Nov 2025
Abstract
The lymphocytic choriomeningitis virus (LCMV) is an under-investigated rodent-borne arenavirus primarily associated with its natural reservoir, the cosmopolitan rodent Mus musculus. Although widely distributed in mice worldwide, human cases are rare, likely under-reported, and often misdiagnosed. While typically asymptomatic or self-limiting, infection
[...] Read more.
The lymphocytic choriomeningitis virus (LCMV) is an under-investigated rodent-borne arenavirus primarily associated with its natural reservoir, the cosmopolitan rodent Mus musculus. Although widely distributed in mice worldwide, human cases are rare, likely under-reported, and often misdiagnosed. While typically asymptomatic or self-limiting, infection can progress to neurological disease, severe congenital outcomes, or fatal illness in transplant recipients. Despite its public health relevance, this study provides the first detection and characterization of LCMV in Brazil. We analyzed 236 rodent serum samples and 78 tissue samples from synanthropic rodents (Mus musculus, Rattus rattus, and Rattus norvegicus) collected during seven independent expeditions across the state of Rio de Janeiro, Southeastern Brazil. Using ELISAs, IgG anti-LCMV antibodies were detected in 20% of rodents, including two R. rattus (2/10), two R. norvegicus (2/95), and forty-five M. musculus (45/131). The LCMV’s RNA was amplified and partially sequenced from fourteen M. musculus, and complete S segment sequences were obtained from two mouse samples. Phylogenetic analyses revealed that these Brazilian strains belong to lineage I, which is composed of strains that induce disease in humans. Our results underscore the importance of implementing integrated surveillance for this zoonosis in Brazil.
Full article
(This article belongs to the Special Issue Rodent-Borne Viruses 2026)
►▼
Show Figures
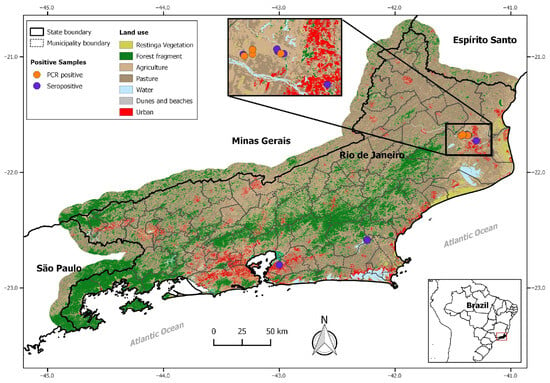
Figure 1
Open AccessArticle
Chronic Chikungunya Arthritis in Northeastern Brazil: An Association with Very Severe Joint Pain and Lack of Correlation with IL-6 and TNFα Gene Polymorphisms
by
Mariella Sousa Coêlho Maciel, Catharina Diniz de Brito Martins, Alan Gleison Moreira dos Santos, Caroline Nobre Oliveira, Hygor Ferreira Fernandes, Raphael de Oliveira Rodrigues and Juliana Navarro Ueda Yaochite
Viruses 2025, 17(12), 1543; https://doi.org/10.3390/v17121543 - 26 Nov 2025
Abstract
Chikungunya fever (CHIKF) affects thousands of people annually. Therefore, aspects involved in the pathogenesis of the disease must be further explored. In this study, we assessed the development of chronic chikungunya arthritis (CCA) and the single-nucleotide polymorphisms (SNPs) -174 G/C in the interleukin-6
[...] Read more.
Chikungunya fever (CHIKF) affects thousands of people annually. Therefore, aspects involved in the pathogenesis of the disease must be further explored. In this study, we assessed the development of chronic chikungunya arthritis (CCA) and the single-nucleotide polymorphisms (SNPs) -174 G/C in the interleukin-6 (IL-6) gene and -308 G/A in the tumor necrosis factor alpha (TNFα) gene on CHIKF development in a population from Northeastern Brazil. A total of 313 blood and serum samples were collected. Of these, 102 were included in the cases group, 182 were included in the control group, and 29 were incorporated in the asymptomatic group. The DNA extraction and genotyping across real-time quantitative polymerase chain reaction (RT-qPCR) were performed. The data showed that 71.6% of individuals who had CHIKF developed CCA. A significant increase in CCA development was observed in females, and very severe joint pain was associated with increased risk of CCA. In contrast, we did not identify significant associations between the SNPs and CHIKF development. Our data indicate that females and individuals who develop very severe joint pain have an increased risk for CCA and highlight the lack of correlation of SNPs in the IL-6 and TNFα genes with CHIKF in the studied population.
Full article
(This article belongs to the Special Issue Innate and Adaptive Immune Responses to Arbovirus Infections)
►▼
Show Figures
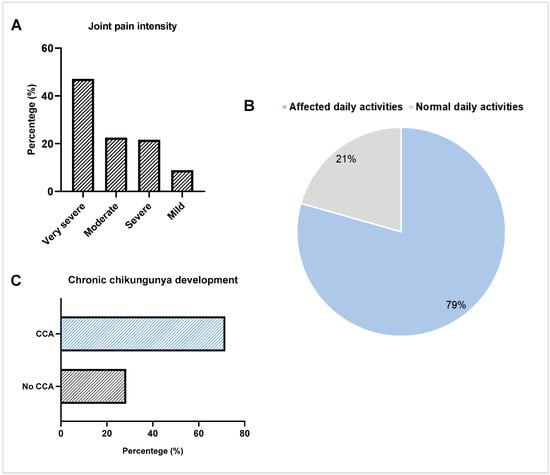
Figure 1
Open AccessArticle
Early-Life Demographic Factors Shape Gut Microbiome Patterns Associated with Rotavirus Gastroenteritis Severity
by
Eman R. Abdelbary, Mohammed Ramadan, Ibrahim A. Amin, Fatma S. Abd-Elsamea, Ashraf Mohamed Elsaghier, Eman Ahmed Abd-Alrahman, Hani A. Ozbak, Hassan A. Hemeg, Yahya A. Almutawif, Shadi A. Zaki, Ali A. Abdelrahman and Mohammed Salah
Viruses 2025, 17(12), 1542; https://doi.org/10.3390/v17121542 - 26 Nov 2025
Abstract
Background: Rotavirus gastroenteritis (RVGE) remains a leading cause of severe infant diarrhea worldwide, with growing evidence supporting the role of the gut microbiome in modulating the disease. However, the interplay between early-life demographic factors, the gut microbiome, and their combined impact on RVGE
[...] Read more.
Background: Rotavirus gastroenteritis (RVGE) remains a leading cause of severe infant diarrhea worldwide, with growing evidence supporting the role of the gut microbiome in modulating the disease. However, the interplay between early-life demographic factors, the gut microbiome, and their combined impact on RVGE clinical severity remains inadequately characterized, particularly in specific geographic populations. Aim: We aimed to investigate how demographic determinants shape gut microbiome composition and function in RVGE and how these features relate to clinical severity. Methods: In our comprehensive case–control study of 165 infants (120 RVGE cases and 45 healthy controls, aged 0–12 months), we utilized 16S rRNA sequencing combined with advanced statistical modeling and machine learning to investigate how demographic factors influence microbiome composition and clinical outcomes. Results: RVGE cases exhibited significantly reduced bacterial diversity (Kruskal–Wallis, Static = 14.85, p < 0.001) and distinct patterns, with community structure most strongly associated with dehydration severity (PERMANOVA; R2 = 0.15, p < 0.001). Substantial taxonomic alterations were identified characterized by depletion of beneficial commensals including Akkermansia (LDA score = 3.8, p < 0.001), Faecalibacterium (Random Forest AUC = 0.82, p < 0.001), and Bifidobacterium (r = −0.42 with breastfeeding, p < 0.001), alongside enrichment of inflammation-associated taxa such as Escherichia-Shigella (WBC; r = 0.49, p < 0.001, and CRP; r = 0.56, p < 0.001), Streptococcus (LDA score = 4.2, p < 0.001), and Staphylococcus. Proteobacteria was the top potential biomarker of severe outcomes (Random Forest AUC = 0.85), with abundance positively correlated with systemic inflammation (CRP: r = 0.51, p = 0.003). Functional predictions revealed increased lipopolysaccharide biosynthesis (ko00540) and reduced butanoate metabolism (ko00650, p < 0.001) in severe disease. Importantly, demographic factors significantly modulated clinical outcomes: cesarean-delivered, formula-fed infants presented the most dysbiotic profiles and experienced 3.2-fold longer hospitalization (95% CI: 1.8–5.6, p < 0.001) than vaginally delivered, breastfed infants did. Conclusions: Collectively, these findings demonstrate that early-life demographic factors potentially shape the gut microbiome composition and function, may influence RVGE severity and recovery trajectories, thus providing candidate biomarkers for risk stratification and identifying targets for microbiota-based interventions.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures
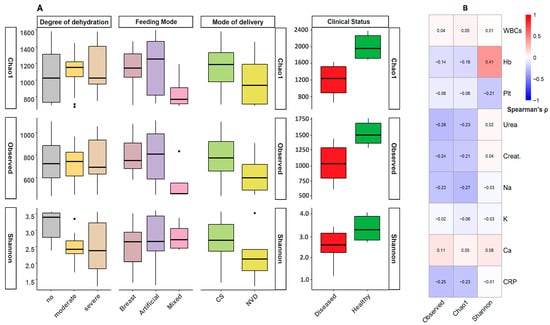
Figure 1
Open AccessArticle
Integrating Machine Learning with Hybrid and Surrogate Models to Accelerate Multiscale Modeling of Acute Respiratory Infections
by
Andrey Korzin, Maria Koshkareva and Vasiliy Leonenko
Viruses 2025, 17(12), 1541; https://doi.org/10.3390/v17121541 - 25 Nov 2025
Abstract
Accurate, efficient, and explainable modeling of the dynamics of acute respiratory infections (ARIs) remains, in many aspects, a significant challenge. While compartmental models such as SIR (Susceptible–Infected–Recovered) remain widely used for that purpose due to their simplicity, they cannot capture the complicated multiscale
[...] Read more.
Accurate, efficient, and explainable modeling of the dynamics of acute respiratory infections (ARIs) remains, in many aspects, a significant challenge. While compartmental models such as SIR (Susceptible–Infected–Recovered) remain widely used for that purpose due to their simplicity, they cannot capture the complicated multiscale nature of disease progression which unites individual-level interactions affecting the initial phase of an outbreak and mass action laws governing the disease transmission in its general phase. Individual-based models (IBMs) offer a detailed representation capable of capturing these transmission nuances but have high computational demands. In this work, we explore hybrid and surrogate approaches to accelerate forecasting of acute respiratory infection dynamics performed via detailed epidemic models. The hybrid approach combines IBMs and compartmental models, dynamically switching between them with the help of statistical and ML-based methods. The surrogate approach, on the other hand, replaces IBM simulations with trained autoencoder approximations. Our results demonstrate that the usage of machine learning techniques and hybrid modeling allows us to obtain a significant speed–up compared to the original individual-based model—up to 1.6–2 times for the hybrid approach and up to
(This article belongs to the Special Issue Multiscale Modeling and Forecasting of COVID-19 and Respiratory Virus Dynamics)
►▼
Show Figures

Figure 1
Open AccessReview
The Host Immune Response to Enterovirus A71 (EV-A71): From Viral Immune Evasion to Immunopathology and Prognostic Biomarkers of Severe Disease—A Narrative Review
by
Anna Andronik, Dawid Lewandowski, Artur Sulik and Kacper Toczylowski
Viruses 2025, 17(12), 1540; https://doi.org/10.3390/v17121540 - 25 Nov 2025
Abstract
Enterovirus A71 (EV-A71) is a critical global pathogen, primarily causing Hand-Foot-and-Mouth Disease (HFMD) but frequently leading to severe neurological complications, including fatal neurogenic pulmonary edema (PE). This review elucidates the complex interplay between viral pathogenesis and the host immune response. EV-A71 utilizes receptors
[...] Read more.
Enterovirus A71 (EV-A71) is a critical global pathogen, primarily causing Hand-Foot-and-Mouth Disease (HFMD) but frequently leading to severe neurological complications, including fatal neurogenic pulmonary edema (PE). This review elucidates the complex interplay between viral pathogenesis and the host immune response. EV-A71 utilizes receptors like SCARB2 and PSGL-1 for entry, while its proteases (2Apro, 3Cpro) efficiently evade innate immunity by cleaving key signaling adaptors (MAVS, TRIF), suppressing Type I IFN response. Critical to disease progression is the age-dependent vulnerability in infants and the subsequent shift toward immunopathology. Severe disease is driven by a systemic cytokine storm and T cell dysregulation, characterized by a loss of control from Treg cells and a profound Th17/Treg imbalance, resulting in high levels of pathogenic cytokines (e.g., IL-17A, IFN-γ). Clinical progression is predicted by specific biomarkers, including Treg depletion, monocyte exhaustion (PD-1/PD-L1), and suppressed regulatory signaling (low cAMP). These findings highlight that effective therapeutic strategies must target host-mediated damage through immunomodulation (e.g., by exploring interventions against key pathogenic axes like IL-6 and IL-1β) and call for the development of next-generation vaccines capable of eliciting balanced cellular immunity to prevent immunopathology.
Full article
(This article belongs to the Special Issue An Update on Enterovirus Research, 2nd Edition)
►▼
Show Figures
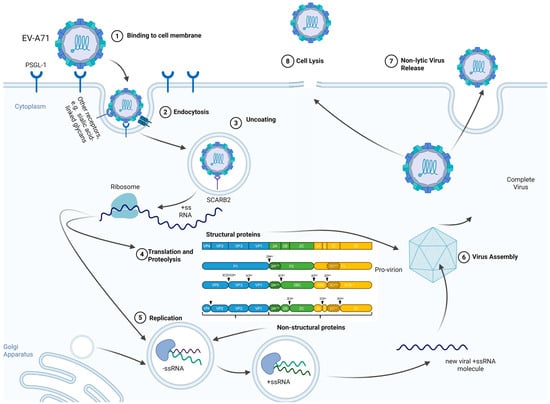
Figure 1
Open AccessArticle
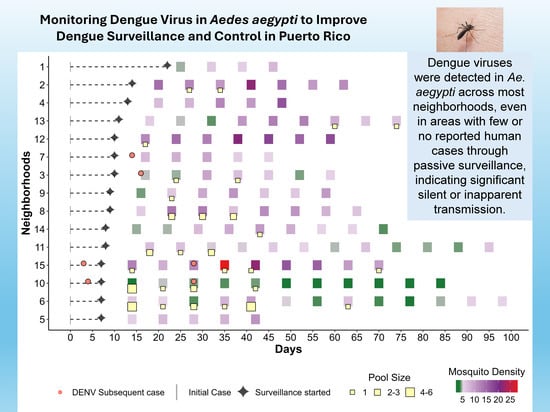
Monitoring Dengue Virus in Aedes aegypti to Improve Dengue Surveillance and Control in Puerto Rico
by
Luisa M. Otero, Joanelis Medina, Jose Ruiz-Valcarcel, Reinaldo Rivera, Yashira Maldonado, Jomil Torres, Zachary J. Madewell, Laura Adams, Gabriela Paz-Bailey and Roberto Barrera
Viruses 2025, 17(12), 1539; https://doi.org/10.3390/v17121539 - 25 Nov 2025
Abstract
Aedes aegypti is the primary urban vector for several important arboviruses, including dengue, chikungunya, yellow fever, and Zika viruses. Traditional dengue virus (DENV) surveillance relies on passive reporting of human cases, which often underestimates transmission due to asymptomatic or unreported infections. This study
[...] Read more.
Aedes aegypti is the primary urban vector for several important arboviruses, including dengue, chikungunya, yellow fever, and Zika viruses. Traditional dengue virus (DENV) surveillance relies on passive reporting of human cases, which often underestimates transmission due to asymptomatic or unreported infections. This study evaluated the utility of monitoring DENV in Ae. aegypti mosquitoes to improve detection of local dengue transmission and inform vector control strategies during the 2024 dengue epidemic in Puerto Rico. Mosquito surveillance was conducted in 15 neighborhoods within the San Juan metropolitan area where confirmed dengue cases had been recently reported. Adult female Ae. aegypti were collected weekly using Autocidal Gravid Ovitraps (AGO traps) placed within a 200 m radius of index cases. Pools of 1–20 mosquitoes were tested for DENV RNA and serotype using RT-PCR. Surveillance continued for up to 91 days in study areas, depending on virus detection. A total of 29,354 female Ae. aegypti were collected, of which 29,211 females were pooled (1–20 specimens per pool) into 3878 pools and analyzed. DENV was detected in 49 pools across 11 neighborhoods, with serotypes DENV-1, DENV-2, and DENV-3 identified. Multiple serotypes were sometimes detected in mosquitoes from the same neighborhood. Minimum infection rates and vector indices were higher during the epidemic than in previous inter-epidemic periods, and mosquito densities exceeded thresholds considered protective against outbreaks. Entomo-virological surveillance detected a greater variety and evenness of serotypes than passive human surveillance. These findings suggest that entomo-virological surveillance can complement passive case surveillance, providing a more comprehensive detection of DENV circulation. Integrating mosquito-based and human surveillance can improve outbreak detection, guide vector control, and aid in reducing dengue burden in affected communities.
Full article
(This article belongs to the Section Invertebrate Viruses)
►▼
Show Figures
Graphical abstract
Open AccessSystematic Review
Bioinformatics Tools and Approaches for Virus Discovery in Genomic Data: A Systematic Review
by
Julia Galeeva, Polina Kuzmichenko, Alexander Manolov, Alexander Lukashev and Elena Ilina
Viruses 2025, 17(12), 1538; https://doi.org/10.3390/v17121538 - 24 Nov 2025
Abstract
The exponential growth of viral metagenomic data has created an urgent need for accurate and scalable tools for virus discovery, yet the extreme diversity, rapid evolution, and limited reference databases for viruses pose unique computational challenges that traditional sequence comparison methods struggle to
[...] Read more.
The exponential growth of viral metagenomic data has created an urgent need for accurate and scalable tools for virus discovery, yet the extreme diversity, rapid evolution, and limited reference databases for viruses pose unique computational challenges that traditional sequence comparison methods struggle to address. This systematic review, conducted in accordance with PRISMA 2020, examines current trends and methodological advances in virus discovery tools from 1990 to 2025. As virus discovery is a broad and multi-dimensional topic, this review focuses on the first-line tools used to analyze the results of high-throughput sequencing. The review was conducted using the PubMed database with a snowballing approach, with over 54 key studies selected for the analysis. These studies encompass the following approaches: alignment-based methods, rapid similarity estimation techniques, profile hidden Markov model methods, combination pipelines, k-mer-based approaches, and machine learning-based methods. The transition from alignment-based to machine learning methods has dramatically improved the detection of divergent viruses, yet challenges remain in interpreting model decisions and handling incomplete viral genomes. This review summarizes current knowledge and potential future directions for the development of virus detection capabilities.
Full article
(This article belongs to the Section General Virology)
►▼
Show Figures
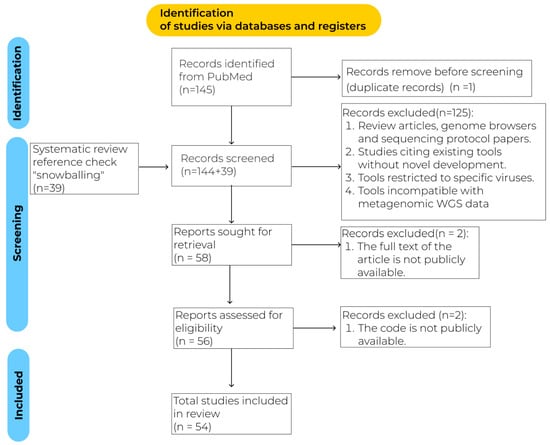
Figure 1
Open AccessArticle
Generation and Characterization of a CE1-Modified mCherry-Expressing Influenza A Virus for In Vivo Imaging and Antiviral Drug Evaluation
by
Zhenghao Li, Meiyi Liu, Jia Yang, Qihui Sun, Dongxue Ye, Wanhui Zhou, Ruikun Du, Shijuan Cheng, Rong Rong, Yong Yang and Xiaoyun Liu
Viruses 2025, 17(12), 1537; https://doi.org/10.3390/v17121537 - 24 Nov 2025
Abstract
Influenza reporter viruses are essential for studying viral infection dynamics and assessing antiviral drug efficacy. However, insertion of exogenous reporter genes can impair both viral replication and reporter expression, limiting the development of these systems. In this study, CE1 compensatory mutation (G3A/
[...] Read more.
Influenza reporter viruses are essential for studying viral infection dynamics and assessing antiviral drug efficacy. However, insertion of exogenous reporter genes can impair both viral replication and reporter expression, limiting the development of these systems. In this study, CE1 compensatory mutation (G3A/C8U) was introduced into the 3′ non-coding region of the NS segment of influenza A/Puerto Rico/8/1934 using reverse genetics, generating the recombinant reporter virus H1N1-PR8-NSCE1-mCherry. Compared with H1N1-PR8-NSWT-mCherry, H1N1-PR8-NSCE1-mCherry produced approximately 2.7-fold more infectious particles. CE1 compensatory mutation partially restored impaired replication kinetics in vitro, as evidenced by higher titers of H1N1-PR8-NSCE1-mCherry at 48 h post-infection in MDCK cells. Additionally, H1N1-PR8-NSCE1-mCherry maintained the intact mCherry gene insertion and high viral titers during serial passaging. Additionally, a real-time, non-invasive in vivo imaging of influenza A viruses was established using H1N1-PR8-NSCE1-mCherry. A significant correlation was observed between lung fluorescence intensity and viral load, indicating that fluorescence signals serve as a reliable indicator of lung viral load in infected mice. Finally, utility of this model for in vivo drug screening was confirmed by antiviral drug oseltamivir phosphate. Above all, H1N1-PR8-NSCE1-mCherry provides a tool for visualizing influenza A virus infection and evaluating antiviral drug efficacy.
Full article
(This article belongs to the Special Issue Antiviral Agents to Influenza Virus 2025)
►▼
Show Figures

Graphical abstract

Journal Menu
► ▼ Journal Menu-
- Viruses Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
Viruses
Infectious Disease Epidemiology and Transmission Dynamics: 3rd Edition
Guest Editors: Zhanwei Du, Lin Wang, Zeynep Ertem, Jose Luis Herrera Diestra, Yuan BaiDeadline: 30 November 2025
Special Issue in
Viruses
Molecular Mechanisms and Clinical Manifestations of Persistent Viral Infections
Guest Editors: Anna Rosa Garbuglia, Simone La FraziaDeadline: 30 November 2025
Special Issue in
Viruses
Research and Clinical Application of Adenovirus (AdV), 3rd Edition
Guest Editor: Julia DavydovaDeadline: 30 November 2025
Special Issue in
Viruses
Advances in Rabies Research 2024
Guest Editors: Charles E. Rupprecht, Juan A. Montaño-HiroseDeadline: 30 November 2025
Topical Collections
Topical Collection in
Viruses
Efficacy and Safety of Antiviral Therapy
Collection Editors: Giordano Madeddu, Andrea De Vito, Agnese Colpani