Antagonistic Trends Between Binding Affinity and Drug-Likeness in SARS-CoV-2 Mpro Inhibitors Revealed by Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Construction and Data Preprocessing
2.2. Computational Environment
2.3. Support Vector Machine (SVM)
2.4. Logistic Regression (LR)
2.5. Molecular Docking and Molecular Dynamics Simulations
3. Results
3.1. Support Vector Machine (SVM) Models
3.2. Logistic Regression (LR) Models
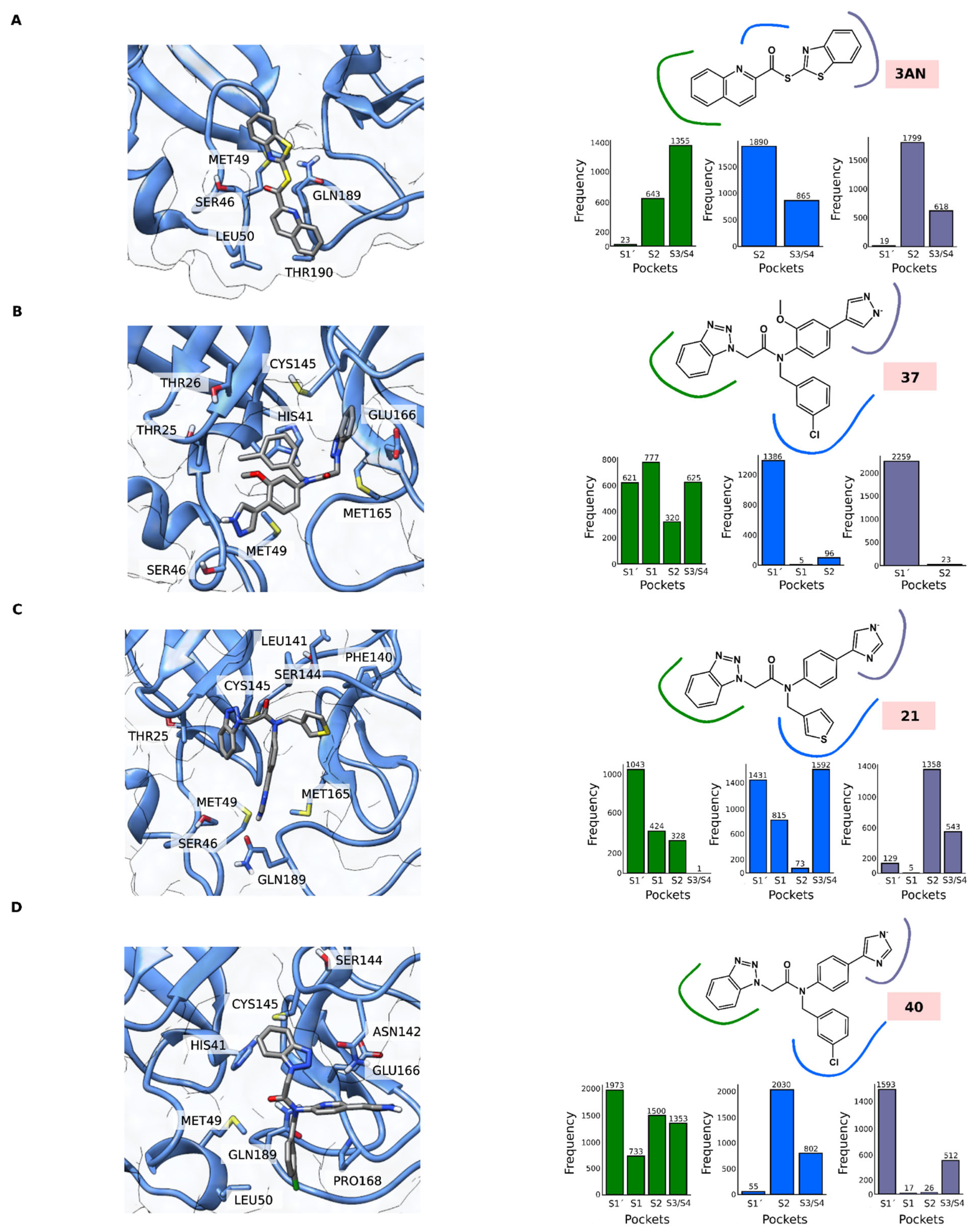
3.3. Molecular Docking and Molecular Dynamics Simulations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Available online: https://covid19.who.int (accessed on 8 June 2025).
- de Souza, A.S.; de Amorim, V.M.F.; Guardia, G.D.A.; dos Santos, F.F.; Ulrich, H.; Galante, P.A.F.; de Souza, R.F.; Guzzo, C.R. Severe Acute Respiratory Syndrome Coronavirus 2 Variants of Concern: A Perspective for Emerging More Transmissible and Vaccine-Resistant Strains. Viruses 2022, 14, 827. [Google Scholar] [CrossRef] [PubMed]
- GISAID Genomic Epidemiology of SARS-CoV-2 with Subsampling Focused Globally over the Past 6 Months. Available online: https://www.gisaid.org/phylodynamics/global/nextstrain/ (accessed on 25 January 2025).
- Chen, J.; Zhang, W.; Li, Y.; Liu, C.; Dong, T.; Chen, H.; Wu, C.; Su, J.; Li, B.; Zhang, W.; et al. Bat-Infecting Merbecovirus HKU5-CoV Lineage 2 Can Use Human ACE2 as a Cell Entry Receptor. Cell 2025, 188, 1729–1742.e16. [Google Scholar] [CrossRef] [PubMed]
- De Amorim, V.M.F.; Soares, E.P.; de Ferrari, A.S.A.; Merighi, D.G.S.; de Souza, R.F.; Guzzo, C.R.; de Souza, A.S. 3-Chymotrypsin-like Protease (3CLpro) of SARS-CoV-2: Validation as a Molecular Target, Proposal of a Novel Catalytic Mechanism, and Inhibitors in Preclinical and Clinical Trials. Viruses 2024, 16, 844. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, Y.; Liu, X.; Jin, Z.; Duan, Y.; Zhang, Q.; Wu, C.; Feng, L.; Du, X.; Zhao, J.; et al. Structural Basis for Replicase Polyprotein Cleavage and Substrate Specificity of Main Protease from SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2022, 119, e2117142119. [Google Scholar] [CrossRef]
- Parmar, M.; Thumar, R.; Patel, B.; Athar, M.; Jha, P.C.; Patel, D. Structural Differences in 3C-like Protease (Mpro) from SARS-CoV and SARS-CoV-2: Molecular Insights Revealed by Molecular Dynamics Simulations. Struct. Chem. 2023, 34, 1309–1326. [Google Scholar] [CrossRef]
- Schechter, I.; Berger, A. On the Size of the Active Site in Proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Boby, M.L.; Fearon, D.; Ferla, M.; Filep, M.; Koekemoer, L.; Robinson, M.C.; COVID Moonshot Consortium; Chodera, J.D.; Lee, A.A.; London, N.; et al. Open Science Discovery of Potent Noncovalent SARS-CoV-2 Main Protease Inhibitors. Science 2023, 382, eabo7201. [Google Scholar] [CrossRef]
- Li, X.; Song, Y. Structure and Function of SARS-CoV and SARS-CoV-2 Main Proteases and Their Inhibition: A Comprehensive Review. Eur. J. Med. Chem. 2023, 260, 115772. [Google Scholar] [CrossRef]
- Noske, G.D.; de Souza Silva, E.; de Godoy, M.O.; Dolci, I.; Fernandes, R.S.; Guido, R.V.C.; Sjö, P.; Oliva, G.; Godoy, A.S. Structural Basis of Nirmatrelvir and Ensitrelvir Activity against Naturally Occurring Polymorphisms of the SARS-CoV-2 Main Protease. J. Biol. Chem. 2023, 299, 103004. [Google Scholar] [CrossRef]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and Calpain Inhibitors II, XII Inhibit SARS-CoV-2 Viral Replication by Targeting the Viral Main Protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef]
- ASAP AI-Driven Structure-Enabled Antiviral Platform (ASAP). Available online: https://asapdiscovery.org/ (accessed on 26 January 2025).
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- The Open Babel Team. Open Babel—The Chemistry Toolbox—Open Babel Openbabel-3-1-1 Documentation. Available online: https://openbabel.org/ (accessed on 26 January 2025).
- Marvin. MarvinSketch. Available online: https://docs.chemaxon.com/display/lts-europium/marvinsketch-downloads.md (accessed on 26 January 2025).
- de Souza, A.S.; de Souza, R.F.; Guzzo, C.R. Quantitative Structure-Activity Relationships, Molecular Docking and Molecular Dynamics Simulations Reveal Drug Repurposing Candidates as Potent SARS-CoV-2 Main Protease Inhibitors. J. Biomol. Struct. Dyn. 2021, 40, 11339–11356. [Google Scholar] [CrossRef] [PubMed]
- NumPy Community. NumPy Reference. Available online: https://numpy.org/community/ (accessed on 26 January 2025).
- The Pandas Development Team. Pandas-Dev/Pandas. Available online: https://github.com/pandas-dev/pandas (accessed on 26 January 2025).
- Landrum, G.; Tosco, P.; Kelley, B.; Rodriguez, R.; Cosgrove, D.; Vianello, R.; Sriniker; Gedeck, P.; Jones, G.; NadineSchneider; et al. Rdkit/rdkit: 2024_09_6 (Q3 2024) Release. Available online: https://zenodo.org/records/14535873 (accessed on 25 January 2025).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Louppe, G.; Prettenhofer, P.; Weiss, R.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Plotly Technologies Inc. Collaborative Data Science Publisher: Plotly Technologies Inc. Place of Publication: Montréal, QC Date of Publication: 2015. Available online: https://plot.ly (accessed on 25 January 2025).
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- The Matplotlib Development Team. Matplotlib: Visualization with Python. Available online: https://matplotlib.org/ (accessed on 25 January 2025).
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Ropp, P.J.; Kaminsky, J.C.; Yablonski, S.; Durrant, J.D. Dimorphite-DL: An Open-Source Program for Enumerating the Ionization States of Drug-like Small Molecules. J. Cheminform. 2019, 11, 14. [Google Scholar] [CrossRef]
- Azlim Khan, A.K.; Ahamed Hassain Malim, N.H. Comparative Studies on Resampling Techniques in Machine Learning and Deep Learning Models for Drug-Target Interaction Prediction. Molecules 2023, 28, 1663. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- GROMACS 2022 Documentation. Available online: https://manual.gromacs.org/2022/index.html (accessed on 25 January 2025).
- GROMACS 2022 Source Code. Available online: https://manual.gromacs.org/2022/download.html (accessed on 25 January 2025).
- Dodda, L.S.; Vilseck, J.Z.; Tirado-Rives, J.; Jorgensen, W.L. 1.14*CM1A-LBCC: Localized Bond-Charge Corrected CM1A Charges for Condensed-Phase Simulations. J. Phys. Chem. B 2017, 121, 3864–3870. [Google Scholar] [CrossRef] [PubMed]
- Dodda, L.S.; Cabeza de Vaca, I.; Tirado-Rives, J.; Jorgensen, W.L. LigParGen Web Server: An Automatic OPLS-AA Parameter Generator for Organic Ligands. Nucleic Acids Res. 2017, 45, W331–W336. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Tirado-Rives, J. Potential Energy Functions for Atomic-Level Simulations of Water and Organic and Biomolecular Systems. Proc. Natl. Acad. Sci. USA 2005, 102, 6665–6670. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Pajouhesh, H.; Lenz, G.R. Medicinal Chemical Properties of Successful Central Nervous System Drugs. NeuroRx 2005, 2, 541–553. [Google Scholar] [CrossRef]
- Bickerton, G.R.; Paolini, G.V.; Besnard, J.; Muresan, S.; Hopkins, A.L. Quantifying the Chemical Beauty of Drugs. Nat. Chem. 2012, 4, 90–98. [Google Scholar] [CrossRef]
- Hitchcock, S.A.; Pennington, L.D. Structure-Brain Exposure Relationships. J. Med. Chem. 2006, 49, 7559–7583. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, J.; Cao, R.; Xu, M.; Wu, Y.; Shang, W.; Wang, X.; Zhang, H.; Jiang, X.; Sun, Y.; et al. Comparative Antiviral Efficacy of Viral Protease Inhibitors against the Novel SARS-CoV-2 In Vitro. Virol. Sin. 2020, 35, 776–784. [Google Scholar] [CrossRef]
- Mahdi, M.; Mótyán, J.A.; Szojka, Z.I.; Golda, M.; Miczi, M.; Tőzsér, J. Analysis of the Efficacy of HIV Protease Inhibitors against SARS-CoV-2’s Main Protease. Virol. J. 2020, 17, 190. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, Y.; Shi, C.; Zhang, Y. Deep Learning-Based Molecular Dynamics Simulation for Structure-Based Drug Design against SARS-CoV-2. Comput. Struct. Biotechnol. J. 2022, 20, 5014–5027. [Google Scholar] [CrossRef] [PubMed]
- Sadybekov, A.V.; Katritch, V. Computational Approaches Streamlining Drug Discovery. Nature 2023, 616, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, A.S.; Reddy, V.P.; Marques, O. Artificial Intelligence and COVID-19: A Multidisciplinary Approach. Integr. Med. Res. 2020, 9, 100434. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.; Harun Ai Rashid, M.; Mridul, M.; Mohanty, C.; Swayamsiddha, S. Application of Artificial Intelligence in COVID-19 Drug Repurposing. Diabetes Metab. Syndr. 2020, 14, 1027–1031. [Google Scholar] [CrossRef]
- Tan, G.S.Q.; Sloan, E.K.; Lambert, P.; Kirkpatrick, C.M.J.; Ilomäki, J. Drug Repurposing Using Real-World Data. Drug Discov. Today 2023, 28, 103422. [Google Scholar] [CrossRef]
- Sharma, J.; Kumar Bhardwaj, V.; Singh, R.; Rajendran, V.; Purohit, R.; Kumar, S. An in-Silico Evaluation of Different Bioactive Molecules of Tea for Their Inhibition Potency against Non Structural Protein-15 of SARS-CoV-2. Food Chem. 2021, 346, 128933. [Google Scholar] [CrossRef]
- Avti, P.; Chauhan, A.; Shekhar, N.; Prajapat, M.; Sarma, P.; Kaur, H.; Bhattacharyya, A.; Kumar, S.; Prakash, A.; Sharma, S.; et al. Computational Basis of SARS-CoV 2 Main Protease Inhibition: An Insight from Molecular Dynamics Simulation Based Findings. J. Biomol. Struct. Dyn. 2021, 40, 8894–8904. [Google Scholar] [CrossRef]
- Gupta, A.; Zhou, H.-X. Profiling SARS-CoV-2 Main Protease (M) Binding to Repurposed Drugs Using Molecular Dynamics Simulations in Classical and Neural Network-Trained Force Fields. ACS Comb. Sci. 2020, 22, 826–832. [Google Scholar] [CrossRef]
- Alves, V.M.; Bobrowski, T.; Melo-Filho, C.C.; Korn, D.; Auerbach, S.; Schmitt, C.; Muratov, E.N.; Tropsha, A. QSAR Modeling of SARS-CoV M Inhibitors Identifies Sufugolix, Cenicriviroc, Proglumetacin, and Other Drugs as Candidates for Repurposing against SARS-CoV-2. Mol. Inform. 2021, 40, e2000113. [Google Scholar] [CrossRef]
- Yu, X. Prediction of Inhibitory Constants of Compounds against SARS-CoV 3CLpro Enzyme with 2D-QSAR Model. J. Saudi Chem. Soc. 2021, 25, 101262. [Google Scholar] [CrossRef]
- Zaki, M.E.A.; Al-Hussain, S.A.; Masand, V.H.; Akasapu, S.; Bajaj, S.O.; El-Sayed, N.N.E.; Ghosh, A.; Lewaa, I. Identification of Anti-SARS-CoV-2 Compounds from Food Using QSAR-Based Virtual Screening, Molecular Docking, and Molecular Dynamics Simulation Analysis. Pharmaceuticals 2021, 14, 357. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Gao, P.; Xu, H.; Liu, Y. Improved SAR and QSAR Models of SARS-CoV-2 Mpro Inhibitors Based on Machine Learning. J. Mol. Liq. 2024, 394, 123708. [Google Scholar] [CrossRef]
- Hazemann, J.; Kimmerlin, T.; Lange, R.; Mac Sweeney, A.; Bourquin, G.; Ritz, D.; Czodrowski, P. Identification of SARS-CoV-2 Mpro Inhibitors through Deep Reinforcement Learning for Drug Design and Computational Chemistry Approaches. RSC Med. Chem. 2024, 15, 2146–2159. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.S.; Martins, J.P.A.; de Melo, E.B. SMILES-Based 2D-QSAR and Similarity Search for Identification of Potential New Scaffolds for Development of SARS-CoV-2 MPRO Inhibitors. Struct. Chem. 2022, 33, 1691–1706. [Google Scholar] [CrossRef]
- Madani, A.; Benkortbi, O.; Laidi, M. In Silico Prediction of the Inhibition of New Molecules on SARS-CoV-2 3CL Protease by Using QSAR: PSOSVR Approach. Braz. J. Chem. Eng. 2023, 41, 427–442. [Google Scholar] [CrossRef]
- Andrianov, A.M.; Shuldau, M.A.; Furs, K.V.; Yushkevich, A.M.; Tuzikov, A.V. AI-Driven De Novo Design and Molecular Modeling for Discovery of Small-Molecule Compounds as Potential Drug Candidates Targeting SARS-CoV-2 Main Protease. Int. J. Mol. Sci. 2023, 24, 8083. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial Intelligence to Deep Learning: Machine Intelligence Approach for Drug Discovery. Mol. Divers. 2021, 25, 1315. [Google Scholar] [CrossRef]
- Sharma, G.; Kumar, N.; Sharma, C.S.; Alqahtani, T.; Tiruneh, Y.K.; Sultana, S.; Rolim Silva, G.V.; de Lima Menezes, G.; Zaki, M.E.A.; Nobre Oliveira, J.I. Identification of Promising SARS-CoV-2 Main Protease Inhibitor through Molecular Docking, Dynamics Simulation, and ADMET Analysis. Sci. Rep. 2025, 15, 2830. [Google Scholar] [CrossRef]
- de Gomes, I.S.; Santana, C.A.; Marcolino, L.S.; de Lima, L.H.F.; de Melo-Minardi, R.C.; Dias, R.S.; de Paula, S.O.; de Silveira, S.A. Computational Prediction of Potential Inhibitors for SARS-CoV-2 Main Protease Based on Machine Learning, Docking, MM-PBSA Calculations, and Metadynamics. PLoS ONE 2022, 17, e0267471. [Google Scholar] [CrossRef]
- Chhetri, S.P.; Bhandari, V.S.; Maharjan, R.; Lamichhane, T.R. Identification of Lead Inhibitors for 3CLpro of SARS-CoV-2 Target Using Machine Learning Based Virtual Screening, ADMET Analysis, Molecular Docking and Molecular Dynamics Simulations. RSC Adv. 2024, 14, 29683–29692. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.N.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An Oral SARS-CoV-2 M Inhibitor Clinical Candidate for the Treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- Blankenship, L.R.; Yang, K.S.; Vulupala, V.R.; Alugubelli, Y.R.; Khatua, K.; Coleman, D.; Ma, X.R.; Sankaran, B.; Cho, C.-C.D.; Ma, Y.; et al. SARS-CoV-2 Main Protease Inhibitors That Leverage Unique Interactions with the Solvent Exposed S3 Site of the Enzyme. ACS Med. Chem. Lett. 2024, 15, 950–957. [Google Scholar] [CrossRef]

| Training Set | Test Set | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Cond. | Split | Non-CV | CV | Non-CV | |||||||||
| Acc. | Rec. | Prec. | F1 | AUC | k = 5 | k = 10 | Acc. | Rec. | Prec. | F1 | AUC | |||
| 1 | 7 | 0.2 | 0.78 | 1.00 | 0.77 | 0.87 | 0.80 | 0.79 | 0.79 | 0.72 | 0.98 | 0.72 | 0.83 | 0.76 |
| 2 | 8 | 0.2 | 0.75 | 1.00 | 0.74 | 0.85 | 0.86 | 0.76 | 0.76 | 0.72 | 1.00 | 0.72 | 0.84 | 0.75 |
| 3 | 9 | 0.2 | 0.79 | 1.00 | 0.78 | 0.88 | 0.86 | 0.79 | 0.80 | 0.72 | 0.98 | 0.72 | 0.83 | 0.80 |
| 4 | 11 | 0.2 | 0.78 | 1.00 | 0.77 | 0.87 | 0.91 | 0.79 | 0.78 | 0.72 | 1.00 | 0.72 | 0.84 | 0.75 |
| 5 | 13 | 0.2 | 0.78 | 1.00 | 0.77 | 0.87 | 0.91 | 0.79 | 0.79 | 0.73 | 1.00 | 0.73 | 0.84 | 0.72 |
| 6 | 40 | 0.2 | 0.81 | 0.72 | 0.76 | 0.74 | 0.89 | 0.82 | 0.82 | 0.80 | 0.70 | 0.74 | 0.72 | 0.85 |
| 7 | 41 | 0.2 | 0.83 | 0.73 | 0.79 | 0.76 | 0.90 | 0.82 | 0.83 | 0.79 | 0.67 | 0.73 | 0.70 | 0.86 |
| 8 | 42 | 0.2 | 0.82 | 0.73 | 0.77 | 0.75 | 0.90 | 0.83 | 0.82 | 0.78 | 0.66 | 0.72 | 0.69 | 0.85 |
| 9 * | 43 | 0.2 | 0.84 | 0.74 | 0.81 | 0.78 | 0.91 | 0.84 | 0.84 | 0.79 | 0.66 | 0.75 | 0.71 | 0.86 |
| 10 | 45 | 0.2 | 0.82 | 0.70 | 0.79 | 0.74 | 0.89 | 0.82 | 0.82 | 0.77 | 0.61 | 0.73 | 0.66 | 0.85 |
| 11 | 22 | 0.25 | 0.72 | 0.63 | 0.70 | 0.66 | 0.22 | 0.72 | 0.72 | 0.71 | 0.63 | 0.67 | 0.65 | 0.23 |
| 12 | 24 | 0.25 | 0.73 | 0.67 | 0.70 | 0.69 | 0.20 | 0.74 | 0.73 | 0.71 | 0.67 | 0.66 | 0.67 | 0.24 |
| 13 | 24 | 0.3 | 0.73 | 0.64 | 0.71 | 0.67 | 0.20 | 0.73 | 0.73 | 0.72 | 0.66 | 0.68 | 0.67 | 0.23 |
| 14 | 29 | 0.2 | 0.80 | 0.71 | 0.80 | 0.76 | 0.12 | 0.80 | 0.78 | 0.72 | 0.63 | 0.70 | 0.66 | 0.22 |
| Training Set | Test Set | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model | Cond. | Solver | Non-CV | k-Fold CV | Non-CV | |||||||||
| Acc. | Rec. | Prec. | F1 | AUC | k = 5 | k = 10 | Acc. | Rec. | Prec. | F1 | AUC | |||
| 1 † | 14 | liblinear | 0.79 | 0.96 | 0.79 | 0.87 | 0.78 | 0.78 | 0.78 | 0.75 | 0.97 | 0.76 | 0.85 | 0.71 |
| 2 † | 14 | saga | 0.79 | 0.96 | 0.79 | 0.87 | 0.78 | 0.78 | 0.79 | 0.75 | 0.97 | 0.76 | 0.85 | 0.70 |
| 3 † | 14 | sag | 0.79 | 0.96 | 0.79 | 0.87 | 0.78 | 0.78 | 0.79 | 0.75 | 0.97 | 0.76 | 0.85 | 0.70 |
| 4 † | 14 | newton-cg | 0.79 | 0.96 | 0.79 | 0.87 | 0.78 | 0.78 | 0.79 | 0.75 | 0.97 | 0.76 | 0.85 | 0.70 |
| 5 † | 14 | lbfgs | 0.79 | 0.96 | 0.79 | 0.87 | 0.78 | 0.78 | 0.79 | 0.75 | 0.97 | 0.76 | 0.85 | 0.70 |
| 6 | 31 | lbfgs | 0.74 | 0.66 | 0.72 | 0.69 | 0.80 | 0.74 | 0.74 | 0.71 | 0.64 | 0.68 | 0.66 | 0.77 |
| 7 | 31 | liblinear | 0.74 | 0.66 | 0.72 | 0.69 | 0.80 | 0.74 | 0.74 | 0.72 | 0.64 | 0.68 | 0.66 | 0.77 |
| 8 | 31 | newton-cg | 0.74 | 0.66 | 0.72 | 0.69 | 0.80 | 0.74 | 0.74 | 0.72 | 0.64 | 0.68 | 0.66 | 0.77 |
| 9 | 31 | sag | 0.74 | 0.66 | 0.72 | 0.69 | 0.80 | 0.74 | 0.73 | 0.71 | 0.64 | 0.68 | 0.66 | 0.76 |
| 10 * | 45 | newton-cg | 0.78 | 0.66 | 0.73 | 0.69 | 0.85 | 0.78 | 0.78 | 0.76 | 0.60 | 0.71 | 0.65 | 0.83 |
| 11 | 45 | sag | 0.78 | 0.65 | 0.72 | 0.69 | 0.85 | 0.78 | 0.78 | 0.76 | 0.60 | 0.71 | 0.65 | 0.83 |
| 12 | 45 | liblinear | 0.78 | 0.66 | 0.73 | 0.69 | 0.85 | 0.78 | 0.78 | 0.76 | 0.60 | 0.71 | 0.65 | 0.83 |
| 13 | 45 | lbfgs | 0.78 | 0.66 | 0.73 | 0.69 | 0.85 | 0.78 | 0.78 | 0.76 | 0.60 | 0.71 | 0.65 | 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, A.S.d.; Amorim, V.M.d.F.; Soares, E.P.; de Souza, R.F.; Guzzo, C.R. Antagonistic Trends Between Binding Affinity and Drug-Likeness in SARS-CoV-2 Mpro Inhibitors Revealed by Machine Learning. Viruses 2025, 17, 935. https://doi.org/10.3390/v17070935
Souza ASd, Amorim VMdF, Soares EP, de Souza RF, Guzzo CR. Antagonistic Trends Between Binding Affinity and Drug-Likeness in SARS-CoV-2 Mpro Inhibitors Revealed by Machine Learning. Viruses. 2025; 17(7):935. https://doi.org/10.3390/v17070935
Chicago/Turabian StyleSouza, Anacleto Silva de, Vitor Martins de Freitas Amorim, Eduardo Pereira Soares, Robson Francisco de Souza, and Cristiane Rodrigues Guzzo. 2025. "Antagonistic Trends Between Binding Affinity and Drug-Likeness in SARS-CoV-2 Mpro Inhibitors Revealed by Machine Learning" Viruses 17, no. 7: 935. https://doi.org/10.3390/v17070935
APA StyleSouza, A. S. d., Amorim, V. M. d. F., Soares, E. P., de Souza, R. F., & Guzzo, C. R. (2025). Antagonistic Trends Between Binding Affinity and Drug-Likeness in SARS-CoV-2 Mpro Inhibitors Revealed by Machine Learning. Viruses, 17(7), 935. https://doi.org/10.3390/v17070935







