Evidence Generation for a Host-Response Biosignature of Respiratory Disease
Abstract
1. Background
2. Methods
2.1. Study Design and Conduct
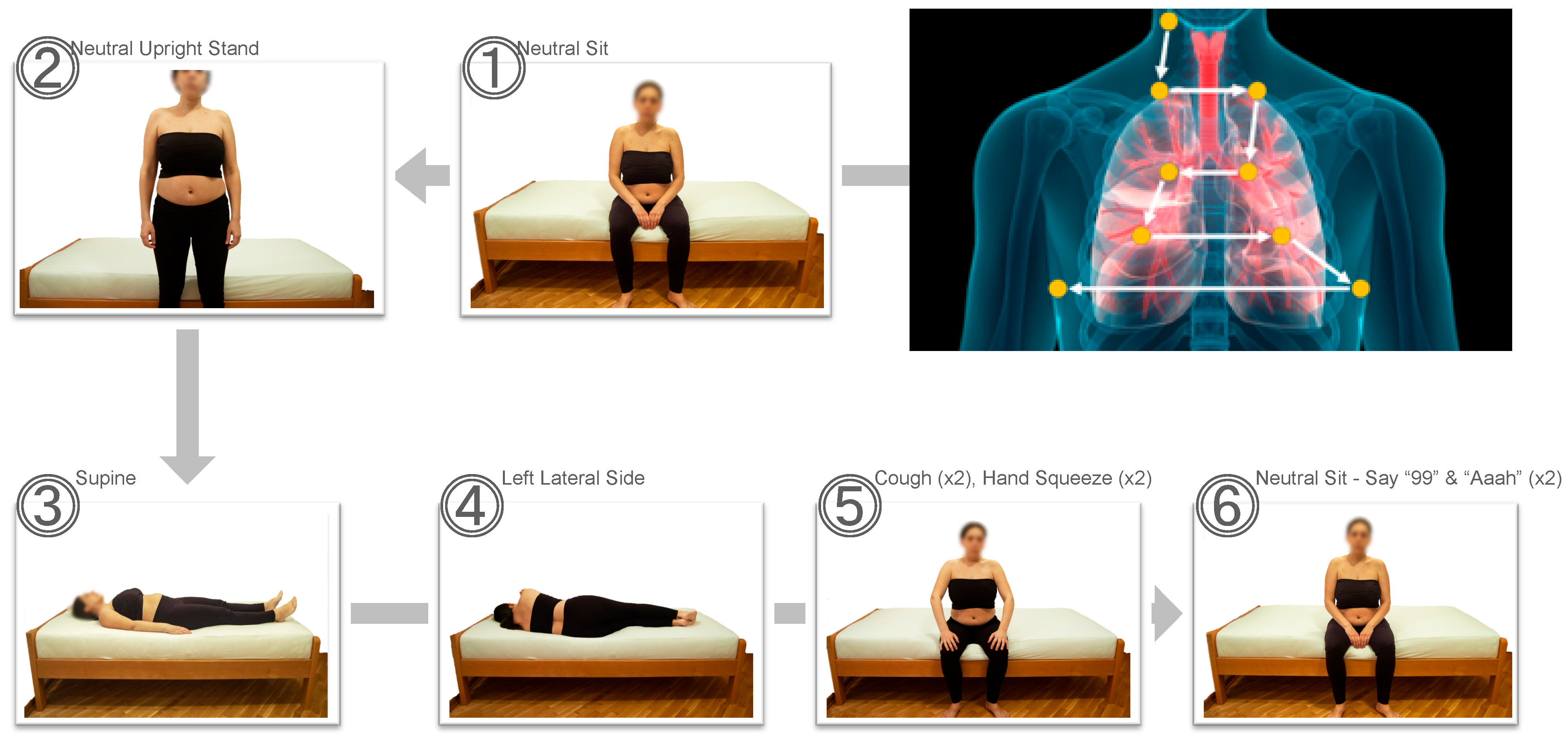
2.2. Study Procedures
2.3. Statistics, Algorithm Training and Testing
3. Results
3.1. Participant Characteristics
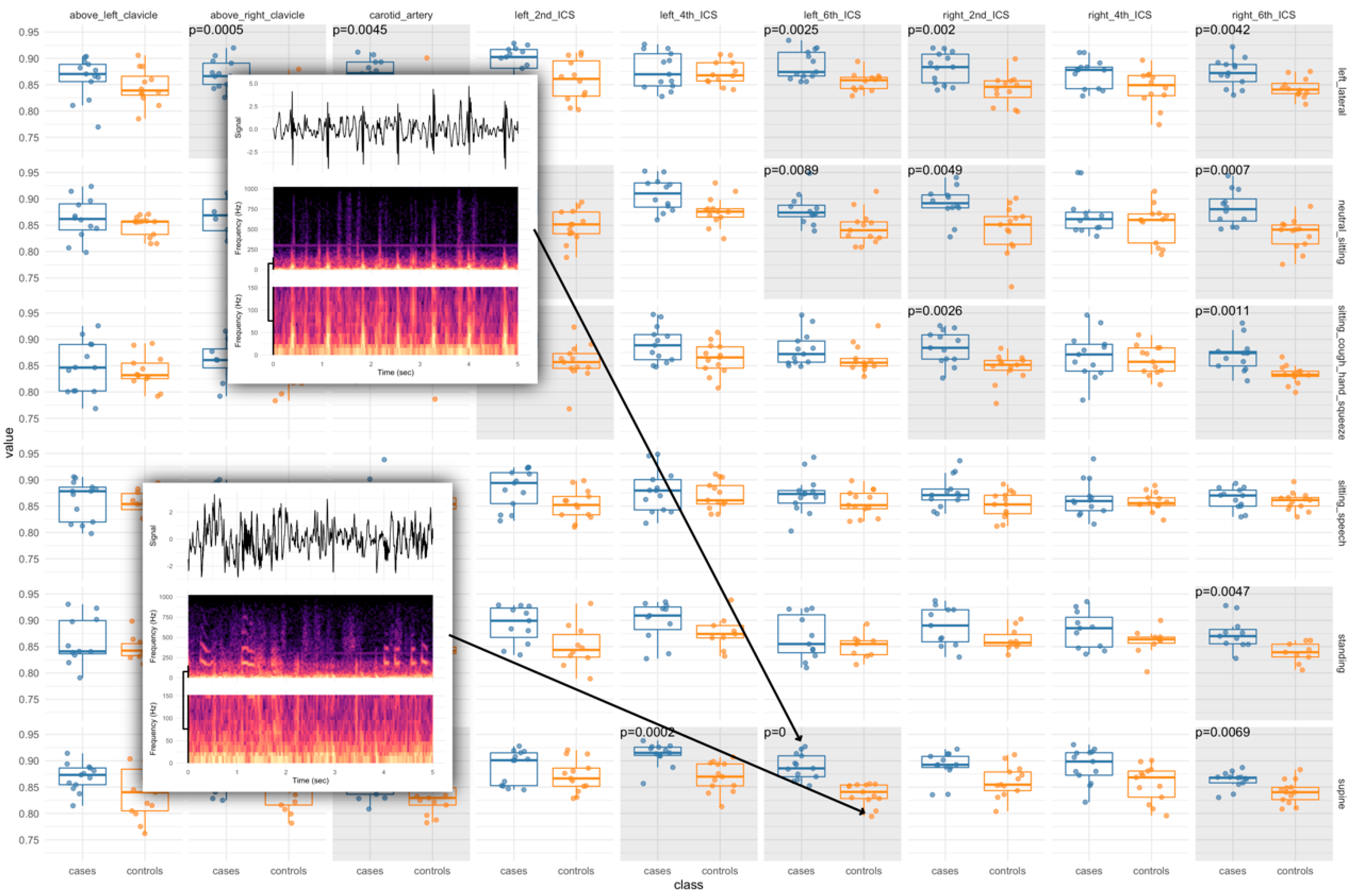
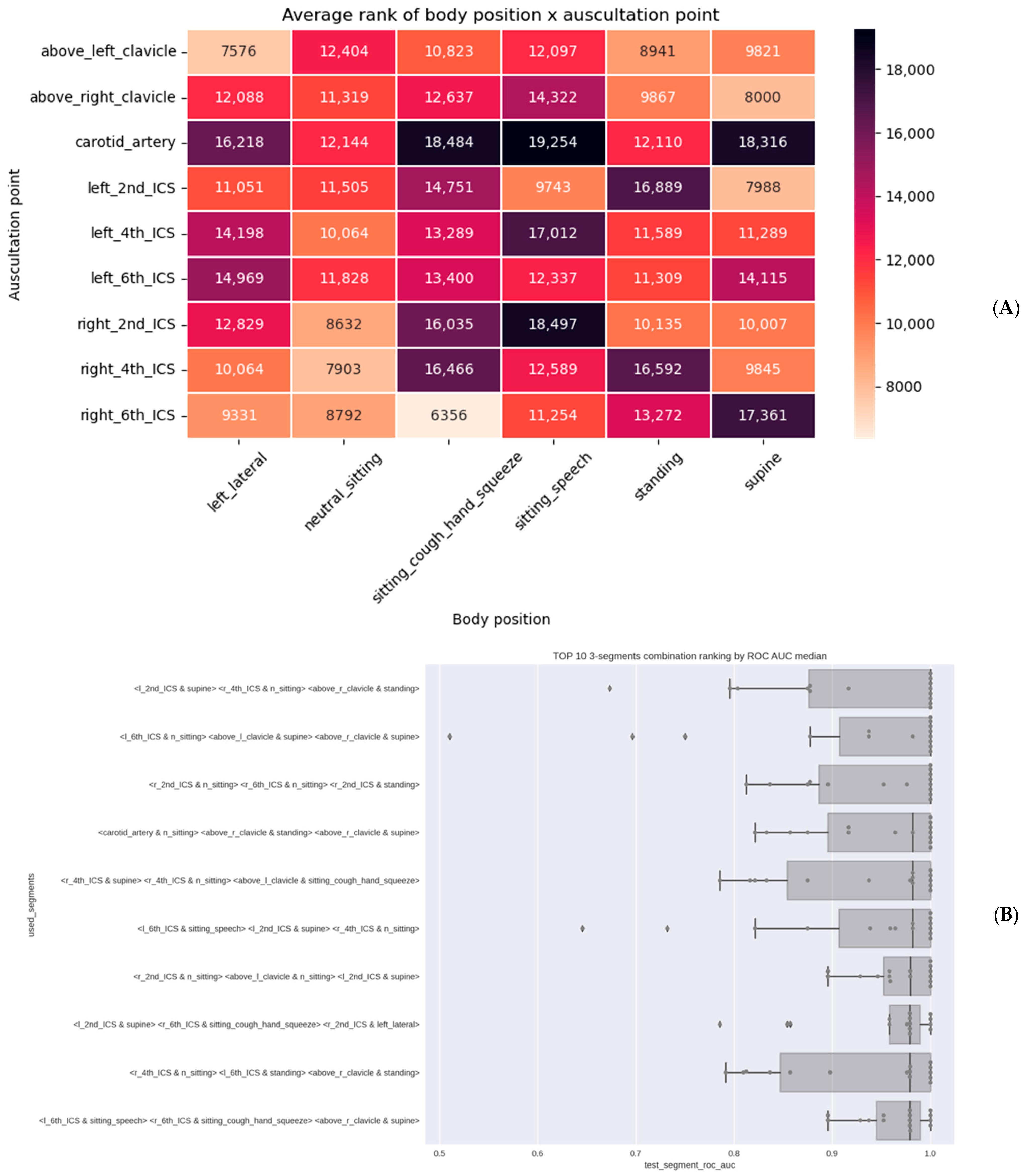
3.2. Algorithm Training and Testing
4. Discussion
4.1. A Biophysics Hypothesis—Directed Respiratory Health Biomarker Testing Framework
4.2. A Rigorous Fast-Fail Matched Case—Control Design for Biomarker Hypothesis Testing
4.3. Establishing High-Quality Ground Truth Data for AI-Based COVID-19 Diagnostics
4.4. A Collaborative-Competition Framework for Curated Crowdsourcing of Cutting-Edge Algorithms
4.5. Study Limitations and Future Steps
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Prior Poster Presentation
References
- İnandıklıoğlu, N.; Akkoc, T. Immune Responses to SARS-CoV, MERS-CoV and SARS-CoV-2. Adv. Exp. Med. Biol. 2020, 1288, 5–12. [Google Scholar] [PubMed]
- Neelakantan, S.; Xin, Y.; Gaver, D.P.; Cereda, M.; Rizi, R.; Smith, B.J.; Avazmohammadi, R. Computational lung modelling in respiratory medicine. J. R. Soc. Interface 2022, 19, 20220062. [Google Scholar] [CrossRef] [PubMed]
- Azarnoosh, J.; Sreenivas, K.; Arabshahi, A.; Newman, J.C. Computational Fluid Dynamics Simulation of the Airflow Through the Human Respiratory Tract. Master’s Thesis, University of Tennessee at Chattanooga, Chattanooga, TN, USA, 2016. [Google Scholar]
- Migulina, N.; Kelley, B.; Zhang, E.Y.; Pabelick, C.M.; Prakash, Y.S.; Vogel, E.R. Mechanosensitive Channels in Lung Health and Disease. Compr. Physiol. 2023, 13, 5157–5178. [Google Scholar] [CrossRef] [PubMed]
- Charras, G.; Yap, A.S. Tensile Forces and Mechanotransduction at Cell–Cell Junctions. Curr. Biol. 2018, 28, R445–R457. [Google Scholar] [CrossRef]
- Cyriac, J.; Huxstep, K. Whistles and Wheezes: Don’t Miss Diseases. Arch. Dis. Child. Educ. Pract. Ed. 2015, 100, 132–143. [Google Scholar] [CrossRef]
- Huan, N.-C.; Sidhu, C.; Thomas, R. Pneumothorax: Classification and Etiology. Clin. Chest Med. 2021, 42, 711–727. [Google Scholar] [CrossRef]
- Hollingsworth, H.M. Wheezing and stridor. Clin. Chest Med. 1987, 8, 231–240. [Google Scholar] [CrossRef]
- Mahler, D.A.; Halpin, D.M.G. Peak Inspiratory Flow as a Predictive Therapeutic Biomarker in COPD. Chest 2021, 160, 491–498. [Google Scholar] [CrossRef]
- Forgacs, P.; Nathoo, A.R.; Richardson, H.D. Breath sounds. Thorax 1971, 26, 288–295. [Google Scholar] [CrossRef]
- Glass, D.S.; Grossfeld, D.; Renna, H.A.; Agarwala, P.; Spiegler, P.; DeLeon, J.; Reiss, A.B. Idiopathic pulmonary fibrosis: Current and future treatment. Clin. Respir. J. 2022, 16, 84–96. [Google Scholar] [CrossRef]
- Volpicelli, G.; Lamorte, A.; Villén, T. What’s new in lung ultrasound during the COVID-19 pandemic. Intensive Care Med. 2020, 46, 1445–1448. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Jain, S.; Miranda, R.; Patil, S.; Pandya, S.; Kotecha, K. Deep learning based respiratory sound analysis for detection of chronic obstructive pulmonary disease. PeerJ Comput. Sci. 2021, 7, e369. [Google Scholar] [CrossRef] [PubMed]
- Sobahi, N.; Atila, O.; Deniz, E.; Sengur, A.; Acharya, U.R. Explainable COVID-19 detection using fractal dimension and vision transformer with Grad-CAM on cough sounds. Biocybern. Biomed. Eng. 2022, 42, 1066–1080. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Qiao, K.; Wang, L.; Yan, B.; Tong, L. Study on Representation Invariances of CNNs and Human Visual Information Processing Based on Data Augmentation. Brain Sci. 2020, 10, 602. [Google Scholar] [CrossRef]
- Cook, A.E.; Garrana, S.H.; Martínez-Jiménez, S.; Rosado-de-Christenson, M.L. Imaging Patterns of Pneumonia. Semin. Roentgenol. 2022, 57, 18–29. [Google Scholar] [CrossRef]
- Haaksma, M.E.; Smit, J.M.; Heldeweg, M.L.A.; Nooitgedacht, J.S.B.; de Grooth, H.J.; Jonkman, A.H.M.; Girbes, A.R.J.; Heunks, L.; Tuinman, P.R. Extended Lung Ultrasound to Differentiate Between Pneumonia and Atelectasis in Critically Ill Patients: A Diagnostic Accuracy Study. Crit. Care Med. 2022, 50, 750–759. [Google Scholar] [CrossRef]
- Jeger-Madiot, N.; Gateau, J.; Fink, M.; Ing, R.-K. Non-contact and through-clothing measurement of the heart rate using ultrasound vibrocardiography. Med. Eng. Phys. 2017, 50, 96–102. [Google Scholar] [CrossRef]
- Lohr, H.A.; van Vollenhoven, E.; van Rotterdam, A. Fundamentals in vibrocardiography. Precordial accelerography and acceleration ballistocardiography. Am. Heart J. 1963, 66, 108–127. [Google Scholar] [CrossRef]
- Will, C.; Shi, K.; Schellenberger, S.; Steigleder, T.; Michler, F.; Fuchs, J.; Weigel, R.; Ostgathe, C.; Koelpin, A. Radar-Based Heart Sound Detection. Sci. Rep. 2018, 8, 11551. [Google Scholar] [CrossRef]
- Abbas, A.K.; Bassam, R. Phonocardiography Signal Processing; Springer International Publishing: Cham, Switzerland, 2009. [Google Scholar] [CrossRef]
- Jaros, R.; Koutny, J.; Ladrova, M.; Martinek, R. Novel phonocardiography system for heartbeat detection from various locations. Sci. Rep. 2023, 13, 14392. [Google Scholar] [CrossRef]
- Agarwal, S.; Mayer, L. Diagnosis and treatment of gastrointestinal disorders in patients with primary immunodeficiency. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2013, 11, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Kaneshiro, M.; Kaiser, W.; Pourmorady, J.; Fleshner, P.; Russell, M.; Zaghiyan, K.; Lin, A.; Martinez, B.; Patel, A.; Nguyen, A.; et al. Postoperative Gastrointestinal Telemetry with an Acoustic Biosensor Predicts Ileus vs. Uneventful GI Recovery. J. Gastrointest. Surg. Off. J. Soc. Surg. Aliment. Tract 2016, 20, 132–139, Discussion 139. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Yadid-Pecht, O.; Sadowski, D.; Mintchev, M.P. Acoustic and Intraluminal Ultrasonic Technologies in the Diagnosis of Diseases in Gastrointestinal Tract: A Review. Engineering 2013, 5, 73–77. [Google Scholar] [CrossRef]
- Pearce, N. Analysis of matched case-control studies. Br. Med. J. 2016, 352, i969. [Google Scholar] [CrossRef] [PubMed]
- Rose, S.; van der Laan, M.J. Why match? Investigating matched case-control study designs with causal effect estimation. Int. J. Biostat. 2009, 5, 1. [Google Scholar] [CrossRef]
- Sousa, A.I.A.D.; Duarte, E.C. Case-control and case-cohort study designs: Methodological considerations. SciELO Prepr. 2023. [Google Scholar] [CrossRef]
- Dinnes, J.; Sharma, P.; Berhane, S.; van Wyk, S.S.; Nyaaba, N.; Domen, J.; Taylor, M.; Cunningham, J.; Davenport, C.; Dittrich, S.; et al. Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2022, 7, CD013705. [Google Scholar]
- Arshadi, M.; Fardsanei, F.; Deihim, B.; Farshadzadeh, Z.; Nikkhahi, F.; Khalili, F.; Sotgiu, G.; Shahidi, B.A.H.; Centis, R.; Migliori, G.B.; et al. Diagnostic Accuracy of Rapid Antigen Tests for COVID-19 Detection: A Systematic Review with Meta-analysis. Front. Med. 2022, 9, 870738. [Google Scholar] [CrossRef]
- Kale, A.U.; Hogg, H.D.J.; Pearson, R.; Glocker, B.; Golder, S.; Coombe, A.; Waring, J.; Liu, X.; Moore, D.J.; Denniston, A.K. Detecting Algorithmic Errors and Patient Harms for AI-Enabled Medical Devices in Randomized Controlled Trials: Protocol for a Systematic Review. JMIR Res. Protoc. 2024, 13, e51614. [Google Scholar] [CrossRef]
- Kaszuba, P.; Turner, A.; Mikulski, B.; Jumbe, N.L.S.; Schuh, A.; Morimoto, M.; Rexelius, P.; Hafen, R.; Deiotte, R.; Hammond, K.; et al. Synthesizing Effective Diagnostic Models from Small Samples using Structural Machine Learning: A Case Study in Automating COVID-19 Diagnosis. In Proceedings of the GECCO’23 Companion: Companion Conference on Genetic and Evolutionary Computation, Lisbon, Portugal, 15–19 July 2023. [Google Scholar]
- Schuh, A.; Morimoto, M.; Kaszuba, P.; Hammond, K.; Swan, J.; Krawiec, K.; Jumbe, N.L.S. Host-directed vibroacoustic biosignature of viral respiratory infection. In Proceedings of the 2023 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI), Pittsburgh, PA, USA, 15–18 October 2023; Sponsored by the IEEE Engineering in Medicine and Biology Society (IEEEEMBS). pp. 1–4. [Google Scholar] [CrossRef]

| All Patients (n = 35) | COVID-19 Negative Group (n = 16) | COVID-19 Positive Group (n = 19) | |
|---|---|---|---|
| Demographic variable | Median (interquartile range) | ||
| Age (years) | 48.6 (30–78) | 49.0 (30–70) | 48.3 (32–78) |
| Body Mass Index (BMI) | 32.4 (19.2–44.3) | 32.4 (19.2–44.3) | 32.4 (21.2–43.6) |
| Demographic variable | N (%) | ||
| Sex | |||
| Male | 15 (43) | 7 (44) | 8 (42) |
| Female | 20 (57) | 9 (56) | 11 (58) |
| Race | |||
| Asian or Asian American | 1 (3) | 0 (0) | 1 (5) |
| Black or African American | 17 (49) | 8 (50) | 9 (47) |
| White | 16 (46) | 8 (50) | 8 (42) |
| Multi race | 1 (3) | 0 (0) | 1 (5) |
| Ethnicity | |||
| Hispanic origin | 3 (9) | 0 (0) | 3 (16) |
| Not of Hispanic origin | 32 (91) | 16 (100) | 16 (84) |
| Smoking history | |||
| Use of tobacco currently or within last 3 months | 3 (9) | 1 (2) | 2 (11) |
| No current tobacco use | 32 (91) | 15 (93) | 17 (89) |
| Laboratory test | Median (interquartile range) | ||
| White blood cell (K/cu mm) | 7.60 (2.21–20.37) | 8.67 (2.59–20.37) | 6.69 (2.21–12.55) |
| Absolute lymphocyte count (K/cu mm) | 1.67 (0.33–5.22) | 2.19 (0.71–5.22) | 1.24 (0.33–3.26) |
| C-Reactive Protein (mg/L) * | 3.88 (0.10–18.50) | 2.04 (0.10–9.30) | 5.42 (0.20–18.50) |
| Erythrocyte sedimentation rate (mm/h) | 51.25 (4.00–130.00) (n = 28) | 54.91 (4.00–130.00) (n = 11) | 48.88 (6.00–88.00) (n = 17) |
| Interleukin 6 (pg/mL) | 34.37 (1.20–372.00) (n = 31) | 20.17 (2.70–100.00) (n = 13) | 44.62 (1.20–372.00) (n = 18) |
| D-dimer (mg/L) | 1.38 (0.28–8.79) (n = 25) | 2.26 (0.31–8.79) (n = 7) | 1.04 (0.28–3.86) (n = 18) |
| Lactate dehydrogenate (U/L) | 260.37 (123.00–576.00) (n = 30) | 220.69 (123.00–339.00) (n = 13) | 290.71 (141.00–576.00) (n = 17) |
| Ferritin (ng/mL) | 833.51 (20.00–10,223.00) | 590.19 (62.00–5286.00) | 1038.42 (20.00–10,223.00) |
| COVID-19 Ab IgG (AU/mL) | 1.40 (0.00–11.90) (n = 30) | 0.51 (0.00–6.59) (n = 13) | 2.08 (0.00–11.90) (n = 17) |
| COVID-19 AB IgA (AU/mL) | 3.18 (0.00–40.04) (n = 30) | 0.00 (0.00–0.00) (n = 13) | 5.62 (0.00–40.04) (n = 17) |
| Medical history | N (%) | ||
| Diabetes | 13 (37) | 8 (50) | 5 (26) |
| Hypertension | 19 (54) | 9 (56) | 10 (53) |
| Hyperlipidemia | 11 (31) | 4 (25) | 7 (37) |
| Coronary artery disease | 3 (9) | 1 (6) | 2 (11) |
| Heart failure | 3 (9) | 2 (13) | 1 (5) |
| Arrhythmia | 1 (3) | 1 (6) | 0 (0) |
| Implantable devices in chest | 2 (6) | 1 (6) | 1 (5) |
| Mechanical heart valve | 0 (0) | 0 (0) | 0 (0) |
| Pacemaker | 0 (0) | 0 (0) | 0 (0) |
| Peripheral artery disease | 1 (3) | 1 (6) | 0 (0) |
| Cerebrovascular disease | 5 (14) | 4 (25) | 1 (5) |
| Chronic lung disease | 4 (11) | 1 (6) | 3 (16) |
| HIV | 4 (11) | 1 (6) | 3 (16) |
| Tuberculosis | 0 (0) | 0 (0) | 0 (0) |
| Liver disease | 2 (6) | 0 (0) | 2 (11) |
| Kidney disease | 7 (20) | 2 (13) | 5 (26) |
| Pregnancy | 1 (3) | 0 (0) | 1 (5) |
| Cancer | 0 (0) | 0 (0) | 0 (0) |
| Dialysis | 0 (0) | 0 (0) | 0 (0) |
| Other immunodeficiency | 3 (9) | 1 (6) | 2 (11) |
| SML1 | SML2 | SML3 # | DL1 | DL2 | |
|---|---|---|---|---|---|
| True Positive | 11 | 13 | 19 | 11 | 10 |
| True Negative | 15 | 14 | 16 | 12 | 17 |
| False Positive | 2 | 3 | 0 | 5 | 0 |
| False Negative | 7 | 5 | 0 | 9 | 8 |
| Accuracy | 74.3% | 77.1% | 100.0% | 62.2% | 77.1% |
| Sensitivity | 61.1% | 72.2% | 100.0% | 55.0% | 55.6% |
| Precision | 84.6% | 81.3% | 100.0% | 68.8% | 100% |
| Specificity | 88.2% | 82.4% | 100.0% | 70.6% | 100% |
| F-score | 71.0% | 76.5% | 100.0% | 61.1% | 71.4% |
| AUC | 78.4% | 83.7% | 100.0% | 64.7% | 75.2% |
| Test Set | AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Reference |
|---|---|---|---|---|
| Temperature assessment | n/a | (0.00–0.23) | (0.90–1.00) | [29] |
| Symptom Assessment | n/a | (0.00–0.60) | (0.66–1.0) | [29] |
| Symptom plus Temp Assessment | n/a | (0.12–0.69) | (0.90–1.00) | [29] |
| Antigen Test vs. symptomatic individuals | n/a | 0.739 (0.684–0.79) | 0.984–0.997 | [30] |
| Antigen Test vs. asymptomatic individuals | n/a | 0.402 (0.215–0.622) | 0.984–0.997 | [30] |
| SML2 (device only) | 0.837 (0.673–0.921) | 0.722 (0.578–0.872) | 0.813 (0.639–0.944) | This manuscript |
| SML3 (device + clinical covariate data) | 1.0 (0.94–1.0) | 1.0 (0.818–1.0) | 0.996 (0.845–1.0) | This manuscript |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dooley, K.E.; Morimoto, M.; Kaszuba, P.; Krasne, M.; Liu, G.; Fuchs, E.; Rexelius, P.; Swan, J.; Krawiec, K.; Hammond, K.; et al. Evidence Generation for a Host-Response Biosignature of Respiratory Disease. Viruses 2025, 17, 943. https://doi.org/10.3390/v17070943
Dooley KE, Morimoto M, Kaszuba P, Krasne M, Liu G, Fuchs E, Rexelius P, Swan J, Krawiec K, Hammond K, et al. Evidence Generation for a Host-Response Biosignature of Respiratory Disease. Viruses. 2025; 17(7):943. https://doi.org/10.3390/v17070943
Chicago/Turabian StyleDooley, Kelly E., Michael Morimoto, Piotr Kaszuba, Margaret Krasne, Gigi Liu, Edward Fuchs, Peter Rexelius, Jerry Swan, Krzysztof Krawiec, Kevin Hammond, and et al. 2025. "Evidence Generation for a Host-Response Biosignature of Respiratory Disease" Viruses 17, no. 7: 943. https://doi.org/10.3390/v17070943
APA StyleDooley, K. E., Morimoto, M., Kaszuba, P., Krasne, M., Liu, G., Fuchs, E., Rexelius, P., Swan, J., Krawiec, K., Hammond, K., Ray, S. C., Hafen, R., Schuh, A., & Jumbe, N. L. S. (2025). Evidence Generation for a Host-Response Biosignature of Respiratory Disease. Viruses, 17(7), 943. https://doi.org/10.3390/v17070943






