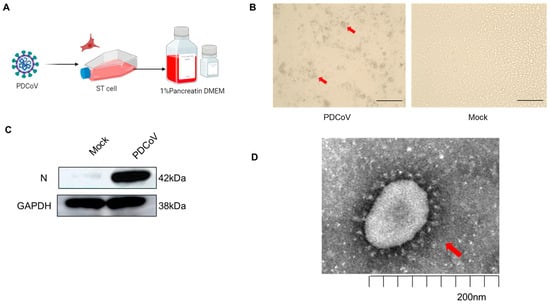
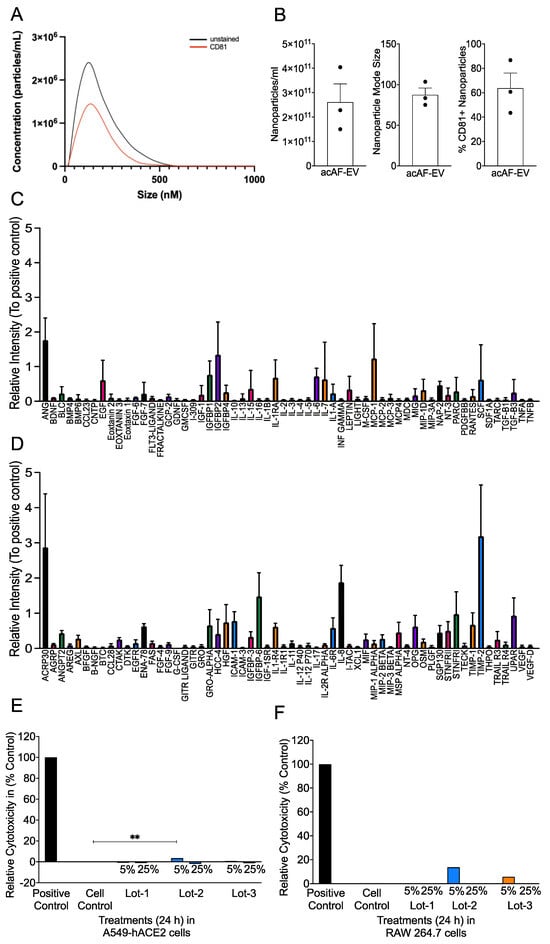
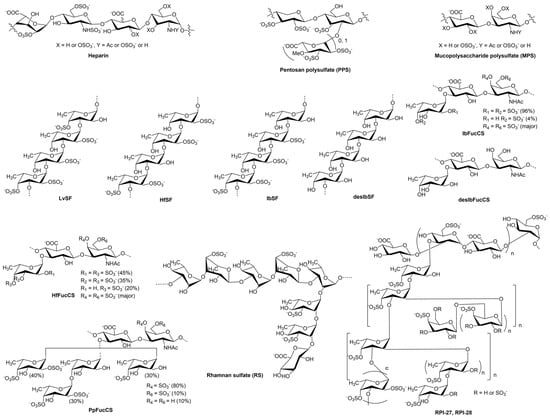
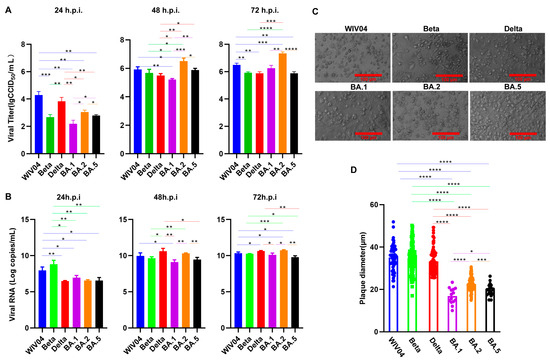
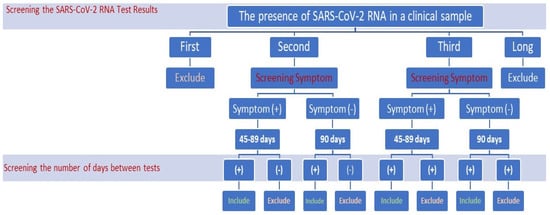
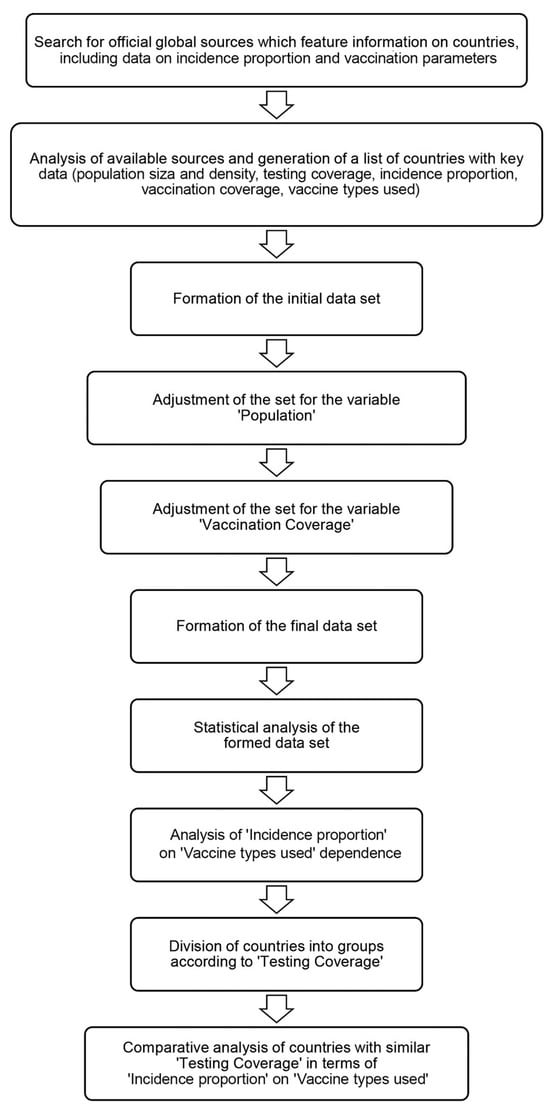
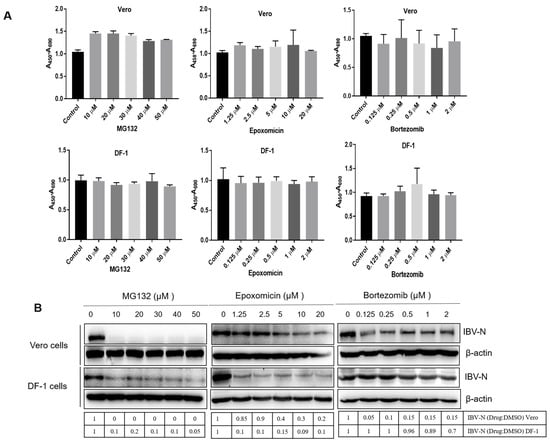
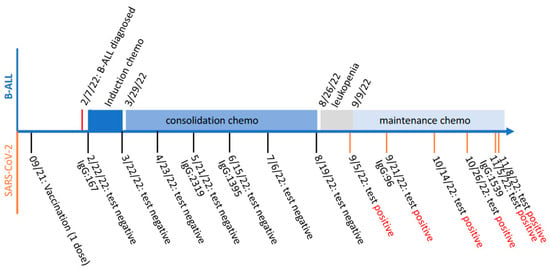
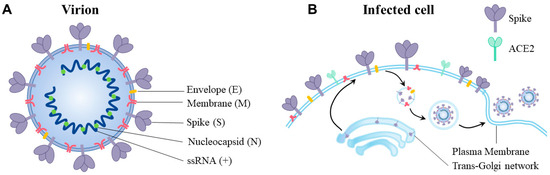
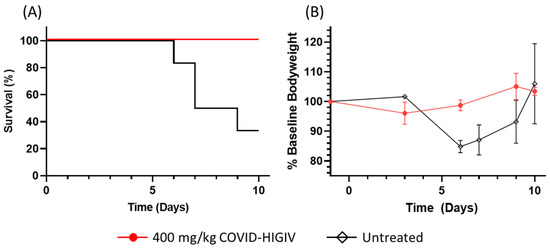
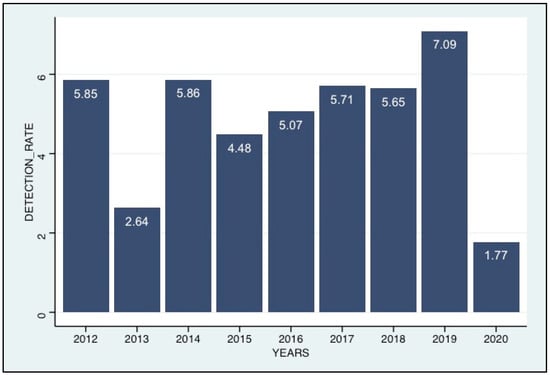
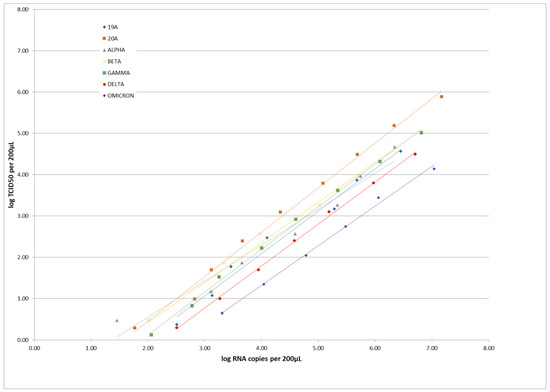
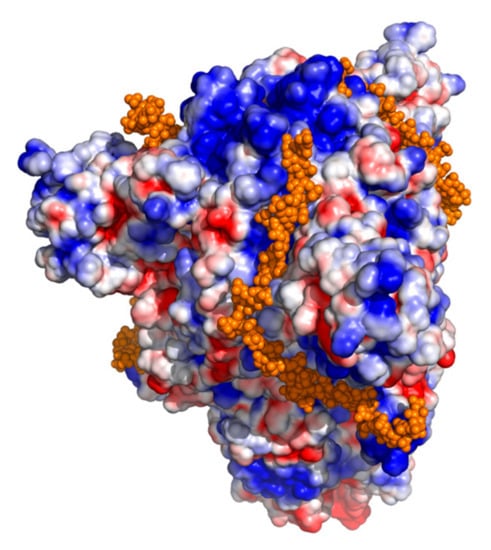
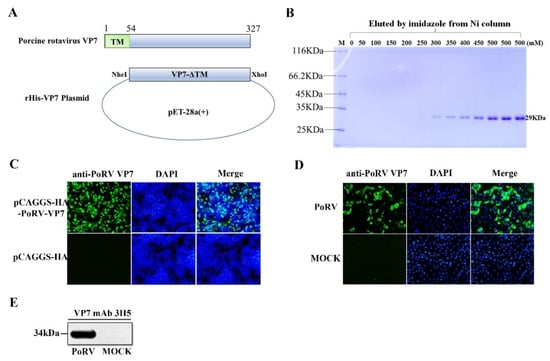
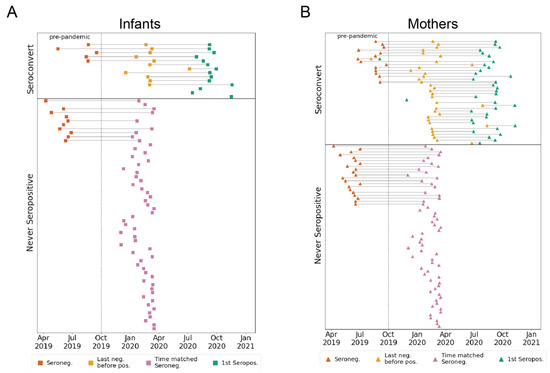
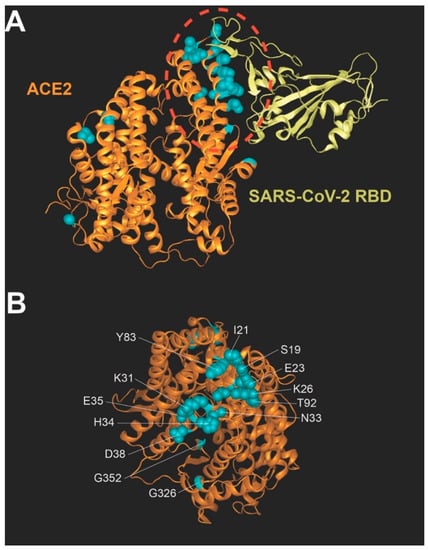
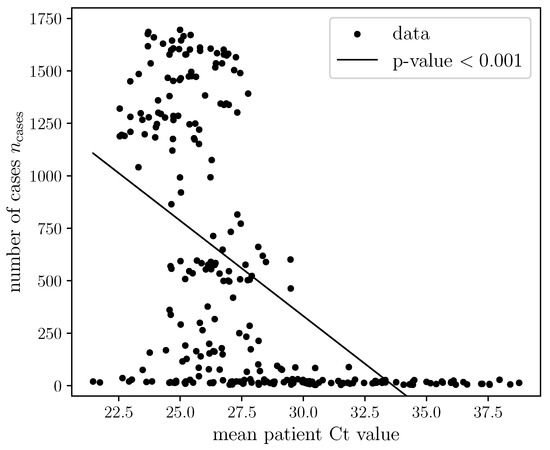
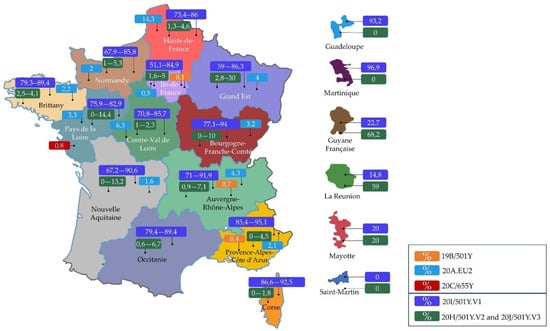
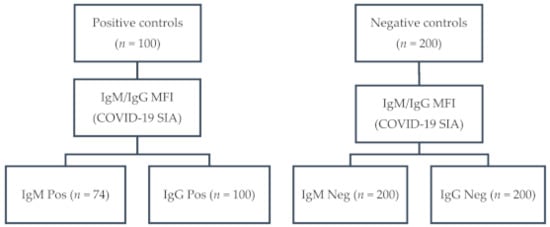
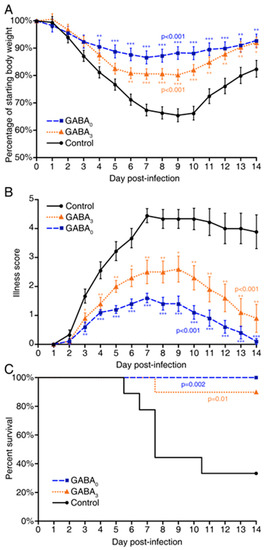
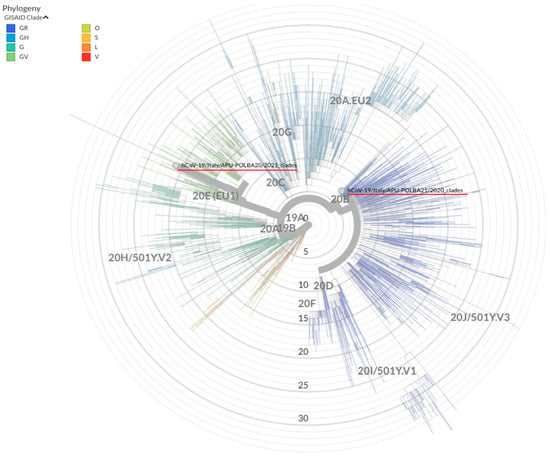
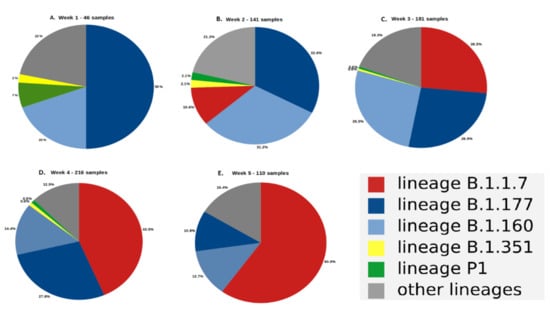
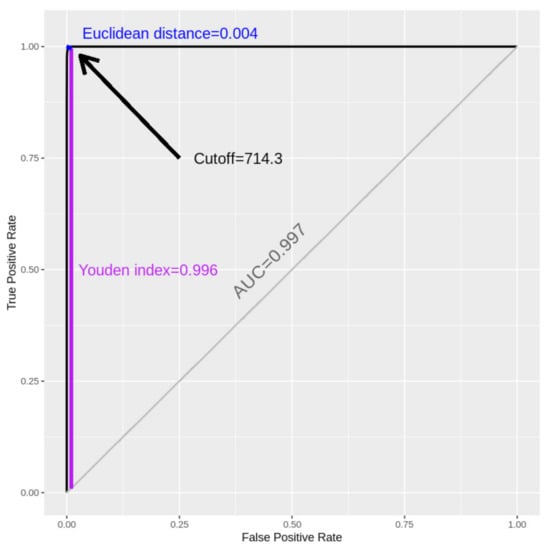
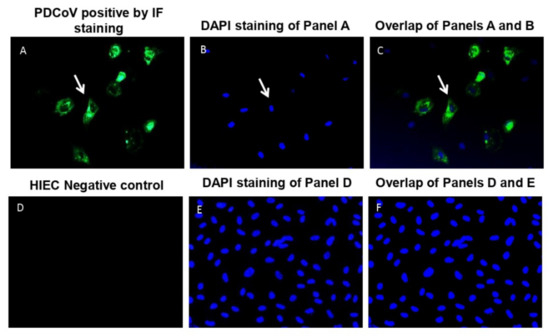
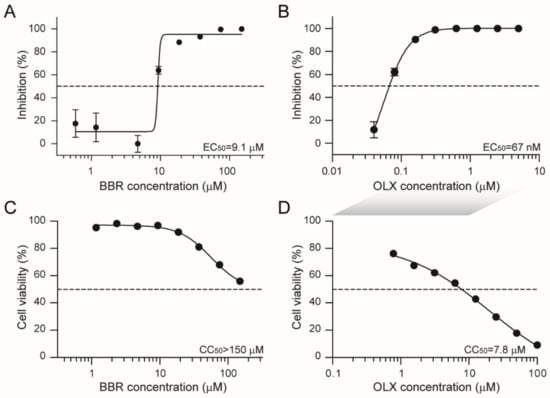
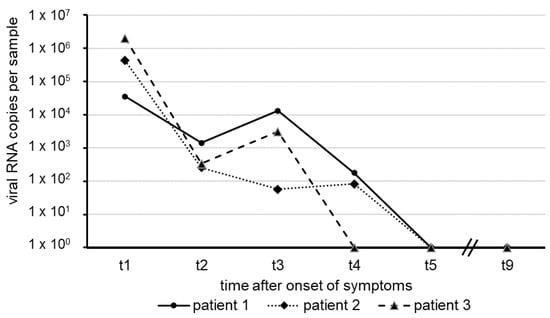
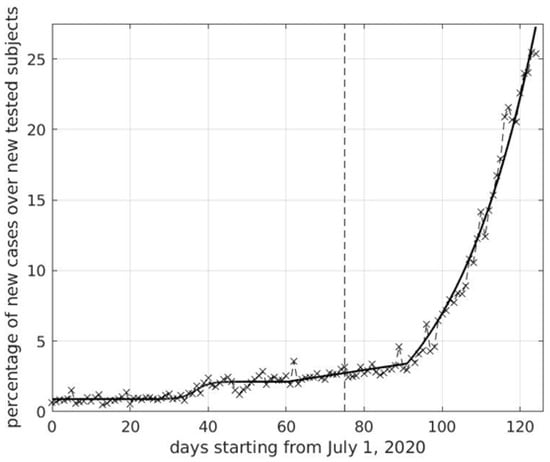
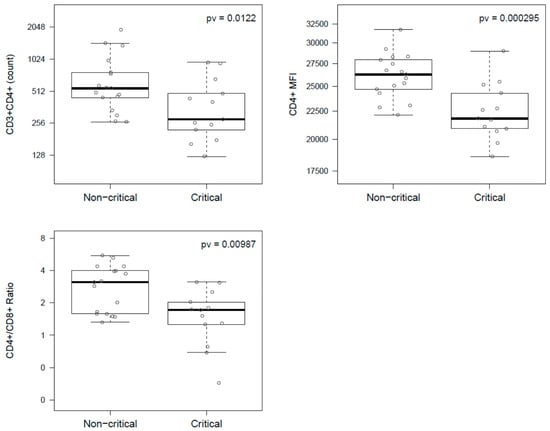
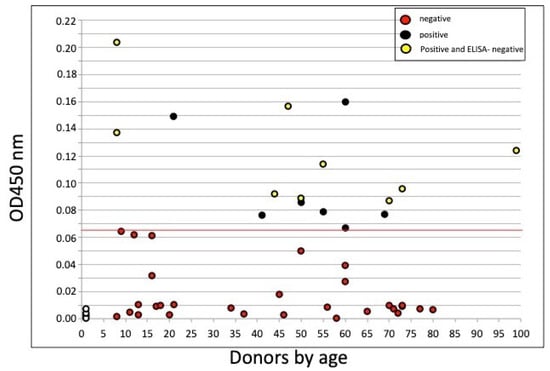
Coronaviruses
A topical collection in Viruses (ISSN 1999-4915). This collection belongs to the section "Coronaviruses".
Submission Status: Closed | Viewed by 564634Editors
Interests: microbiology; virus; antivirals; vaccines; innate immunity; adaptive immunity; interferon
Special Issues, Collections and Topics in MDPI journals
Interests: virology; virus-host interaction; coronavirus; vaccines; antivirals; flavivirus; Zika virus
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
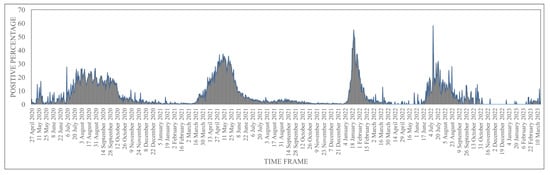
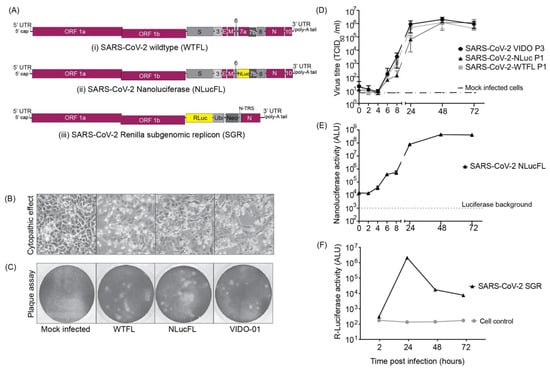
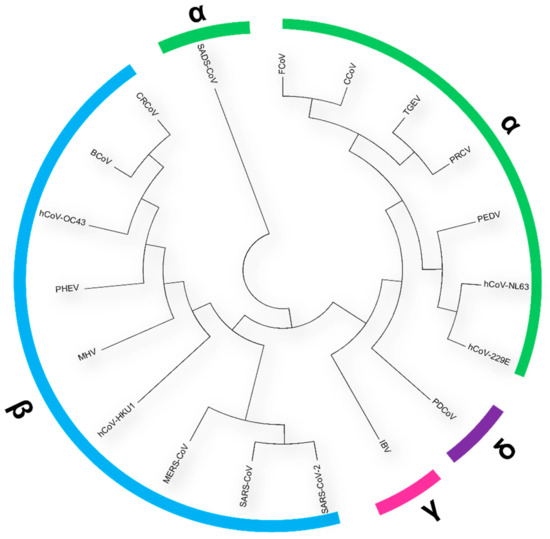
Human coronaviruses (HCoVs), including NL63, 229E, OC43, and HKU1, are enveloped, single-stranded, positive-sense RNA viruses responsible for seasonal mild and moderate respiratory illness (229E and OC43) and, in some cases, bronchiolitis and pneumonia (NL63 and HKU1). However, two HCoVs (SARS and MERS) have been shown to cause severe morbidity and mortality in humans. Recently (December 2019), a novel HCoV, severe acute respiratory syndrome CoV-2 (SARS-CoV-2), was identified in the city of Wuhan in China, and has been responsible for the COVID-19 pandemic, with an alarming case fatality rate, posing an unprecedented threat to human health and the economy. To date, three vaccines have been approved by the Food and Drug Administration (FDA) for emergency use. However, efforts to develop effective countermeasures to deal with SARS-CoV-2 and other HCoV infections are an urgent need.
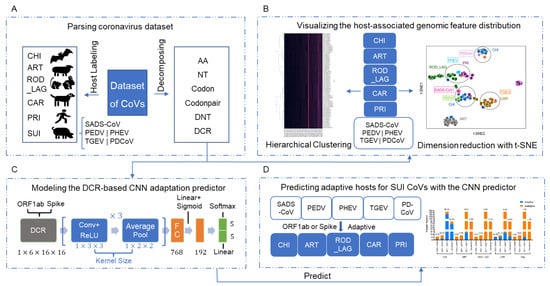
In this Topical Collection about HCoVs, we aim to cover all the aspects related to coronavirus disease not related to SARS-CoV-2 infection and/or COVID-19, including virus–host interactions, viral infection, transmission, pathogenesis, animal models, antivirals, vaccine development, neutralizing antibodies, immunity, innate and adaptive immune responses, prophylactics, therapeutics, and reverse genetics.
We hope that the collection of research articles in this Topical Collection will provide researchers with a better understanding of HCoV infections, with the final goal of using this information for the development of prophylactic and/or therapeutic approaches to controlling these important human respiratory pathogens.
Prof. Dr. Luis Martinez-Sobrido
Dr. Fernando Almazan Toral
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Viruses is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- coronavirus
- HCoV-OC43
- HCoV-229E
- HCoV-NL63
- HCoV-HKU1
- SARS-CoV
- MERS-CoV
- reverse genetics
- immunity
- innate immunity
- adaptive immunity
- virus-host interactions
- antivirals/therapeutics
- vaccines/prophylactics
- inactivated vaccines
- live-attenuated vaccines
- reporter viruses
- animal models
Related Special Issues
- Perspectives and Challenges in Coronavirus Research in Viruses (7 articles)
- Animal Arteriviruses and Coronaviruses in Viruses (17 articles)
- Pathogenesis of Human and Animal Coronaviruses in Viruses (37 articles)
- MERS-CoV in Viruses (17 articles)
- Vaccines and Therapeutics against Coronaviruses in Viruses (27 articles)
- Strategies for the Discovery of Antivirals against Respiratory RNA Viruses in Viruses (5 articles)
- State-of-the-Art Respiratory Viruses Research in Russia in Viruses (18 articles)
- State-of-the-Art Emerging Respiratory Viruses in Europe in Viruses (6 articles)
- SARS-CoV-2 Host Cell Interactions in Viruses (23 articles)
- HIV and SARS-CoV-2 Pathogenesis and Vaccine Development in Viruses (20 articles)
- Co-infections and Superinfections in the Respiratory Tract in Viruses (2 articles)