Porcine Deltacoronavirus-like Particles Produced by a Single Recombinant Baculovirus Elicit Virus-Specific Immune Responses in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, Cells, and Viruses
2.2. Construction of the MSE Triple Expression Plasmid
2.3. Generation of Recombinant Baculovirus
2.4. Indirect Immunofluorescence Assay
2.5. Production and Purification of VLPs
2.6. Western Blotting Analysis
2.7. Transmission Electron Microscopy
2.8. Immunization of Mice
2.9. Determination of IgG Antibodies
2.10. Detection of Neutralizing Antibodies
2.11. Cytokine Release Assay
2.12. Statistical Analysis
3. Results
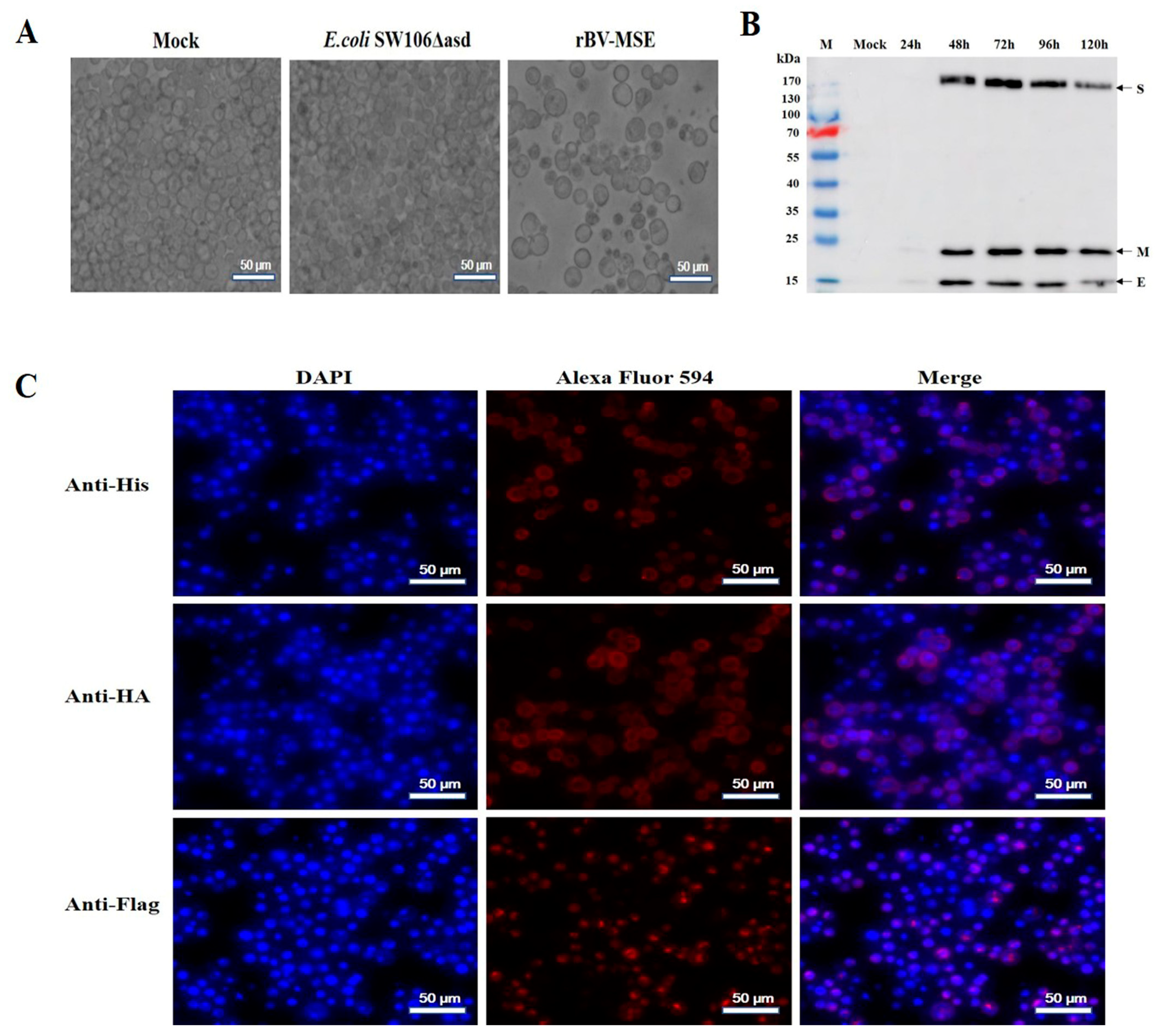
3.1. Generation and Identification of Recombinant Bacmids
3.2. Expression and Analysis of the Recombinant Protein in Sf9 Insect Cells
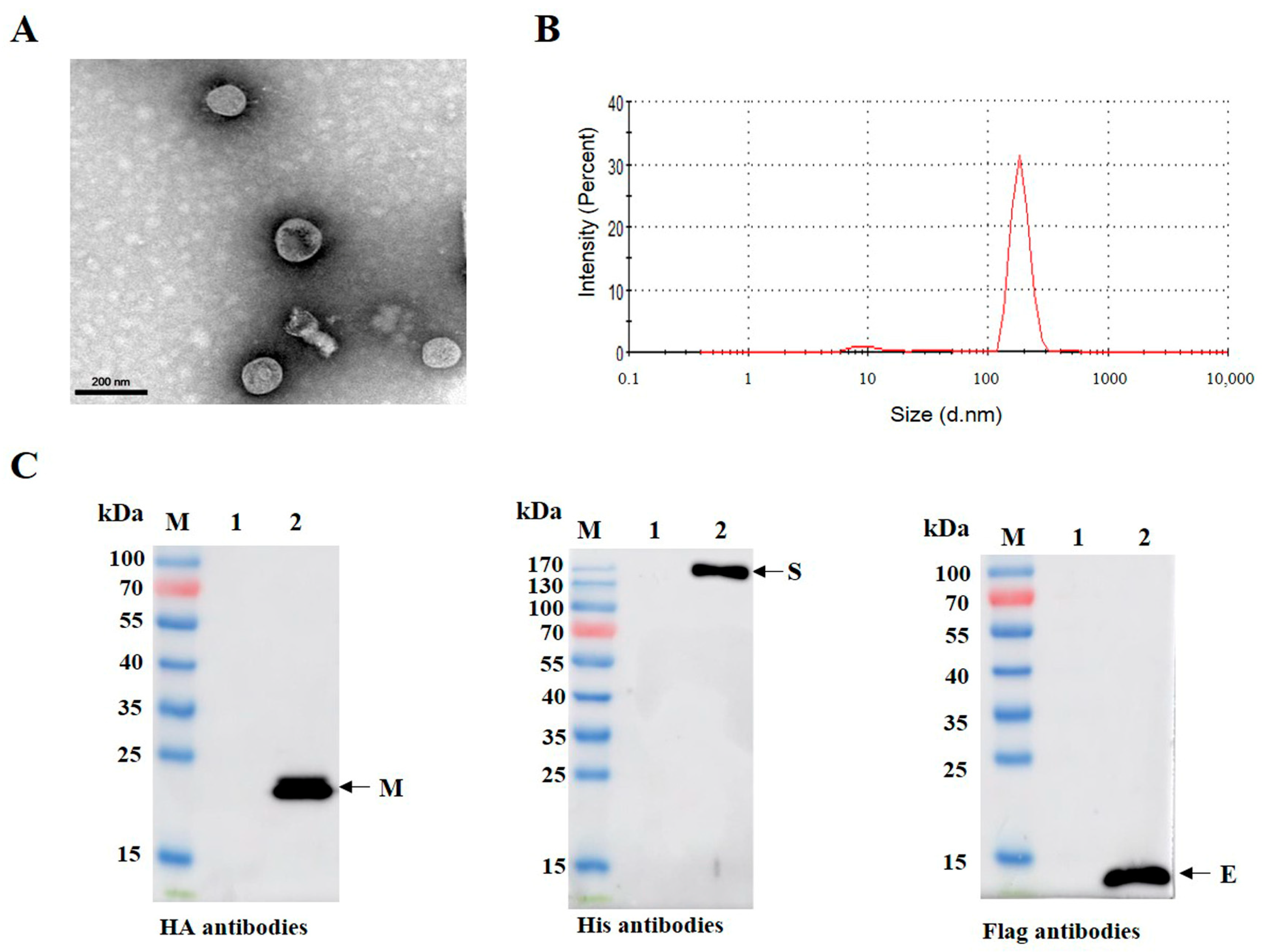
3.3. Purification and Characterization of PDCoV VLPs
3.4. Determination of PDCoV-Specific IgG and NAbs in Mice
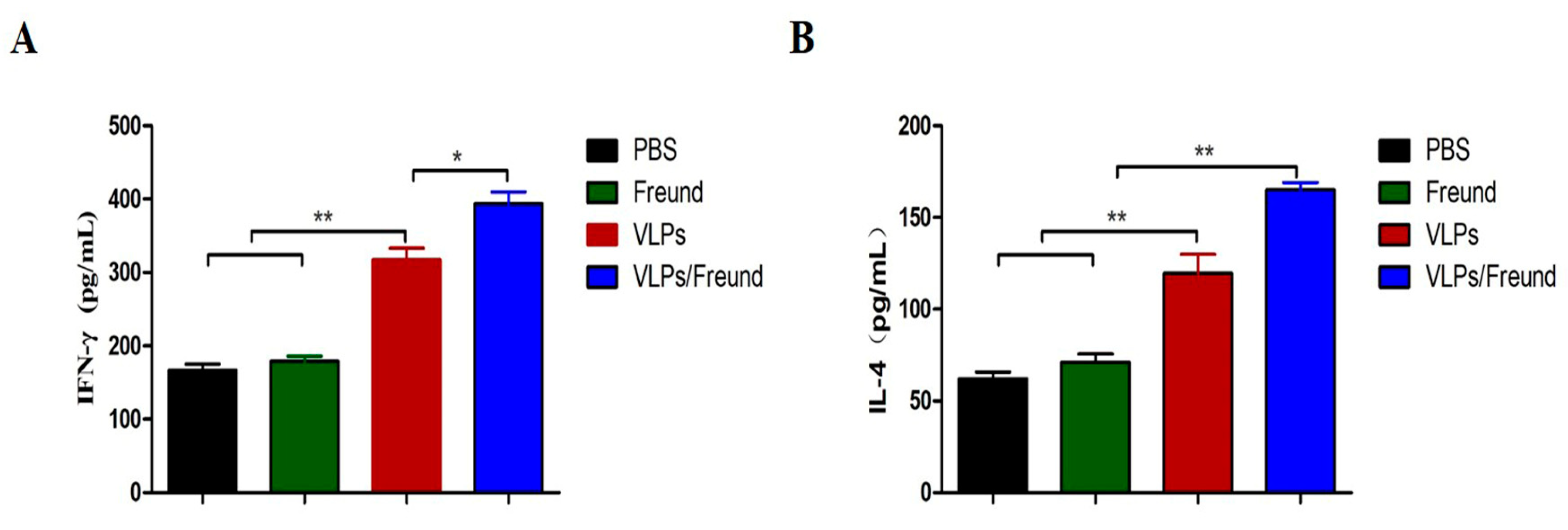
3.5. Analysis of Cytokine Production in Splenocytes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, J. Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016, 226, 71–84. [Google Scholar] [CrossRef]
- Chen, Q.; Gauger, P.; Stafne, M.; Thomas, J.; Arruda, P.; Burrough, E.; Madson, D.; Brodie, J.; Magstadt, D.; Derscheid, R.; et al. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 2015, 482, 51–59. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [PubMed]
- Wang, L.; Byrum, B.; Zhang, Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230. [Google Scholar] [CrossRef]
- Dong, N.; Fang, L.; Zeng, S.; Sun, Q.; Chen, H.; Xiao, S. Porcine deltacoronavirus in mainland China. Emerg. Infect. Dis. 2015, 21, 2254–2255. [Google Scholar] [CrossRef] [PubMed]
- Saeng-Chuto, K.; Lorsirigool, A.; Temeeyasen, G.; Vui, D.T.; Stott, C.J.; Madapong, A.; Tripipat, T.; Wegner, M.; Intrakamhaeng, M.; Chongcharoen, W.; et al. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound. Emerg. Dis. 2017, 64, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Shibahara, T.; Imai, N.; Yamamoto, T.; Ohashi, S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 2018, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States. Emerg. Infect. Dis. 2020, 26, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Hu, H.; Saif, L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017, 162, 2357–2362. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Tagliamonte, M.; Buonaguro, F.M.; Tornesello, M.L.; Buonaguro, L. Virus-like particles as preventive and therapeutic cancer vaccines. Vaccines 2022, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-like particles: Revolutionary platforms for developing vaccines against emerging infectious diseases. Front. Microbiol. 2022, 12, 790121. [Google Scholar] [CrossRef]
- Moradi Vahdat, M.; Hemmati, F.; Ghorbani, A.; Rutkowska, D.; Afsharifar, A.; Eskandari, M.H.; Rezaei, N.; Niazi, A. Hepatitis B core-based virus-like particles: A platform for vaccine development in plants. Biotechnol. Rep. 2021, 29, e00605. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, Y.; Du, J. Human papillomavirus vaccines: An updated review. Vaccines 2020, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Bing, Z.; Guan, S.; Zhang, Z.; Wang, X. Development of new hepatitis E vaccines. Hum. Vaccin. Immunother. 2018, 14, 2254–2262. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, Y.; Xin, G.; Zhou, X.; Lu, H.; Zhang, X.; Xia, X.; Sun, H. Comparative evaluation of ELPylated virus-like particle vaccine with two commercial PCV2 vaccines by experimental challenge. Vaccine 2020, 38, 3952–3959. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoon, J.; Park, J.E. Construction of porcine epidemic diarrhea virus-like particles and its immunogenicity in mice. Vaccines 2021, 9, 370. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, E.; Sung, P.Y.; Wu, W.; Berry, N.; Kempster, S.; Ferguson, D.; Almond, N.; Jones, I.M.; Roy, P. SARS-CoV-2 virus-like particles produced by a single recombinant baculovirus generate anti-S antibody and protect against variant challenge. Viruses 2022, 14, 914. [Google Scholar] [CrossRef]
- Xu, P.W.; Wu, X.; Wang, H.N.; Ma, B.C.; Ding, M.D.; Yang, X. Assembly and immunogenicity of baculovirus-derived infectious bronchitis virus-like particles carrying membrane, envelope and the recombinant spike proteins. Biotechnol. Lett. 2016, 38, 299–304. [Google Scholar] [CrossRef]
- Li, G.; Chen, Q.; Harmon, K.M.; Yoon, K.J.; Schwartz, K.J.; Hoogland, M.J.; Gauger, P.C.; Main, R.G.; Zhang, J. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2014, 2, e00278-14. [Google Scholar] [CrossRef]
- Corse, E.; Machamer, C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 2003, 312, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.G.; Sun, J.C.; Xu, H.; Kan, Y.C.; Zhang, X.; Yan, H. A novel economic method for high throughput production of recombinant baculovirus by infecting insect cells with Bacmid-containing diminopimelate-auxotrophic Escherichia coli. J. Biotechnol. 2010, 145, 23–29. [Google Scholar] [CrossRef]
- Dong, N.; Fang, L.; Yang, H.; Liu, H.; Du, T.; Fang, P.; Wang, D.; Chen, H.; Xiao, S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet. Microbiol. 2016, 196, 98–106. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, J.; Liu, Y.; Da, S.; Shi, H.; Zhang, X.; Liu, J.; Cao, L.; Zhu, X.; Wang, X.; et al. Pathogenicity of porcine deltacoronavirus (PDCoV) strain NH and immunization of pregnant sows with an inactivated PDCoV vaccine protects 5-day-old neonatal piglets from virulent challenge. Transbound. Emerg. Dis. 2020, 67, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, W.; Zhou, P.; Liu, D.; Luo, R.; Jongkaewwattana, A.; He, Q. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: A novel strategy for vaccine design. Emerg. Microbes. Infect. 2020, 9, 20–31. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, Z.; Gu, S.; Nie, M.; Wang, Y.; Zhao, J.; Li, F.; Deng, H.; Huang, J.; Sun, X.; et al. The recombinant pseudorabies virus expressing porcine deltacoronavirus spike protein is safe and effective for mice. BMC Vet. Res. 2022, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zhao, Z.; Huang, Y.W.; Wang, B. Review: A systematic review of virus-like particles of coronavirus: Assembly, generation, chimerism and their application in basic research and in the clinic. Int. J. Biol. Macromol. 2022, 200, 487–497. [Google Scholar] [CrossRef]
- Gopal, R.; Schneemann, A. Production and application of insect virus-based VLPs. Methods Mol. Biol. 2018, 1776, 125–141. [Google Scholar]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Mohsen, M.O.; Gomes, A.C.; Vogel, M.; Bachmann, M.F. Interaction of viral capsid-derived virus-like particles (VLPs) with the innate immune system. Vaccines 2018, 6, 37. [Google Scholar] [CrossRef]
- Schwarz, K.; Meijerink, E.; Speiser, D.E.; Tissot, A.C.; Cielens, I.; Renhof, R.; Dishlers, A.; Pumpens, P.; Bachmann, M.F. Efficient homologous prime-boost strategies for T cell vaccination based on virus-like particles. Eur. J. Immunol. 2005, 35, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.M.; Liu, Y.V.; Mu, H.; Taylor, J.K.; Massare, M.; Flyer, D.C.; Smith, G.E.; Frieman, M.B. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine 2014, 32, 3169–3174. [Google Scholar] [CrossRef]
- Langel, S.N.; Wang, Q.; Vlasova, A.N.; Saif, L.J. Host factors affecting generation of immunity against porcine epidemic diarrhea virus in pregnant and lactating swine and passive protection of neonates. Pathogens 2020, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Bohl, E.H.; Saif, L.J. Passive immunity in transmissible gastroenteritis of swine: Immunoglobulin characteristics of antibodies in milk after inoculating virus by different routes. Infect Immun. 1975, 11, 23–32. [Google Scholar] [CrossRef] [PubMed]


| Primers | Sequence (5′-3′) | PCR Product Size |
|---|---|---|
| MF | ATGTCCGATGCTGAGGAGTGG | 651 bp |
| MR | CATGTACTTATACAGTCGAG | |
| SF | ATGCAACGAGCTTTGTTAAT | 3477 bp |
| SR | CCATTCTTTGAACTTAAAGGAC | |
| EF | ATGGTAGTCGACGACTGGGCC | 249 bp |
| ER | CACGTAATGCGTGTTCCTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Han, X.; Qiao, Y.; Wang, T.; Yao, L. Porcine Deltacoronavirus-like Particles Produced by a Single Recombinant Baculovirus Elicit Virus-Specific Immune Responses in Mice. Viruses 2023, 15, 1095. https://doi.org/10.3390/v15051095
Liu Y, Han X, Qiao Y, Wang T, Yao L. Porcine Deltacoronavirus-like Particles Produced by a Single Recombinant Baculovirus Elicit Virus-Specific Immune Responses in Mice. Viruses. 2023; 15(5):1095. https://doi.org/10.3390/v15051095
Chicago/Turabian StyleLiu, Yangkun, Xueying Han, Yaqi Qiao, Tiejun Wang, and Lunguang Yao. 2023. "Porcine Deltacoronavirus-like Particles Produced by a Single Recombinant Baculovirus Elicit Virus-Specific Immune Responses in Mice" Viruses 15, no. 5: 1095. https://doi.org/10.3390/v15051095
APA StyleLiu, Y., Han, X., Qiao, Y., Wang, T., & Yao, L. (2023). Porcine Deltacoronavirus-like Particles Produced by a Single Recombinant Baculovirus Elicit Virus-Specific Immune Responses in Mice. Viruses, 15(5), 1095. https://doi.org/10.3390/v15051095






