Risk Assessment of the Possible Intermediate Host Role of Pigs for Coronaviruses with a Deep Learning Predictor
Abstract
1. Introduction
2. Methods
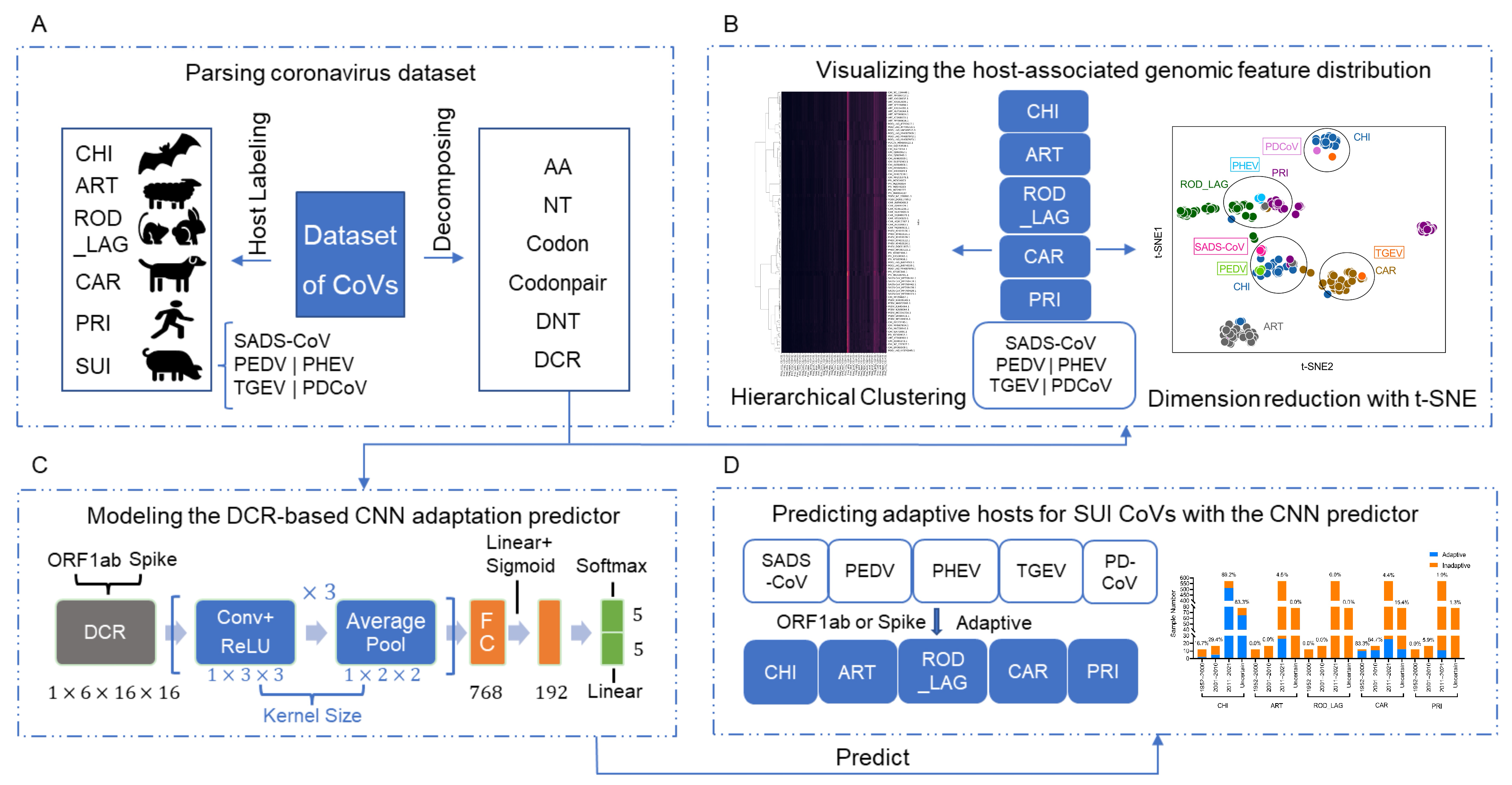
2.1. Data Processing, Host Labeling, and Sequence Decomposition of CoVs
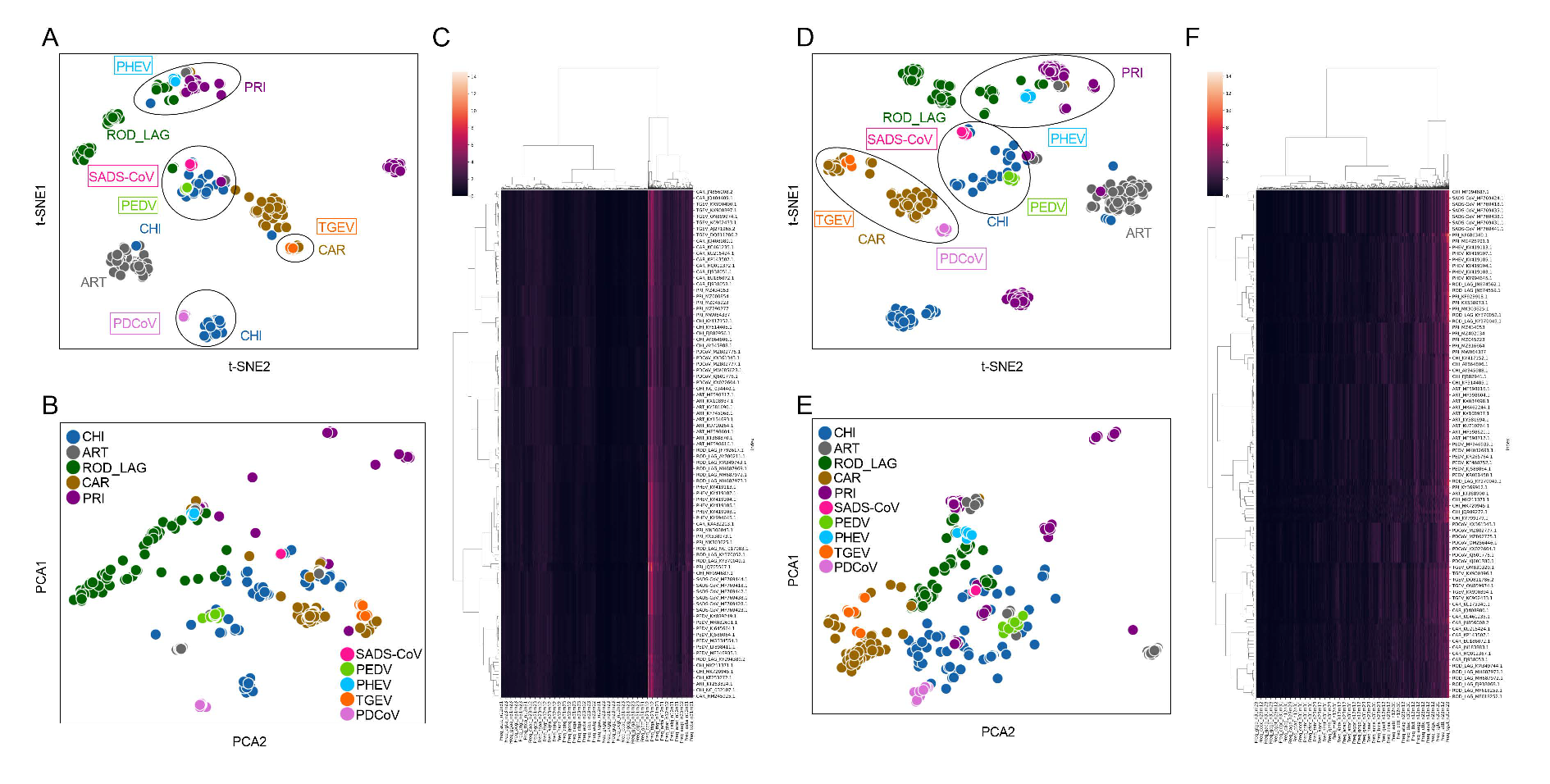
2.2. Reduction, Visualization, and Clustering of Six Types of Compositional Traits of ORF1ab and Spike Sequences of CoVs
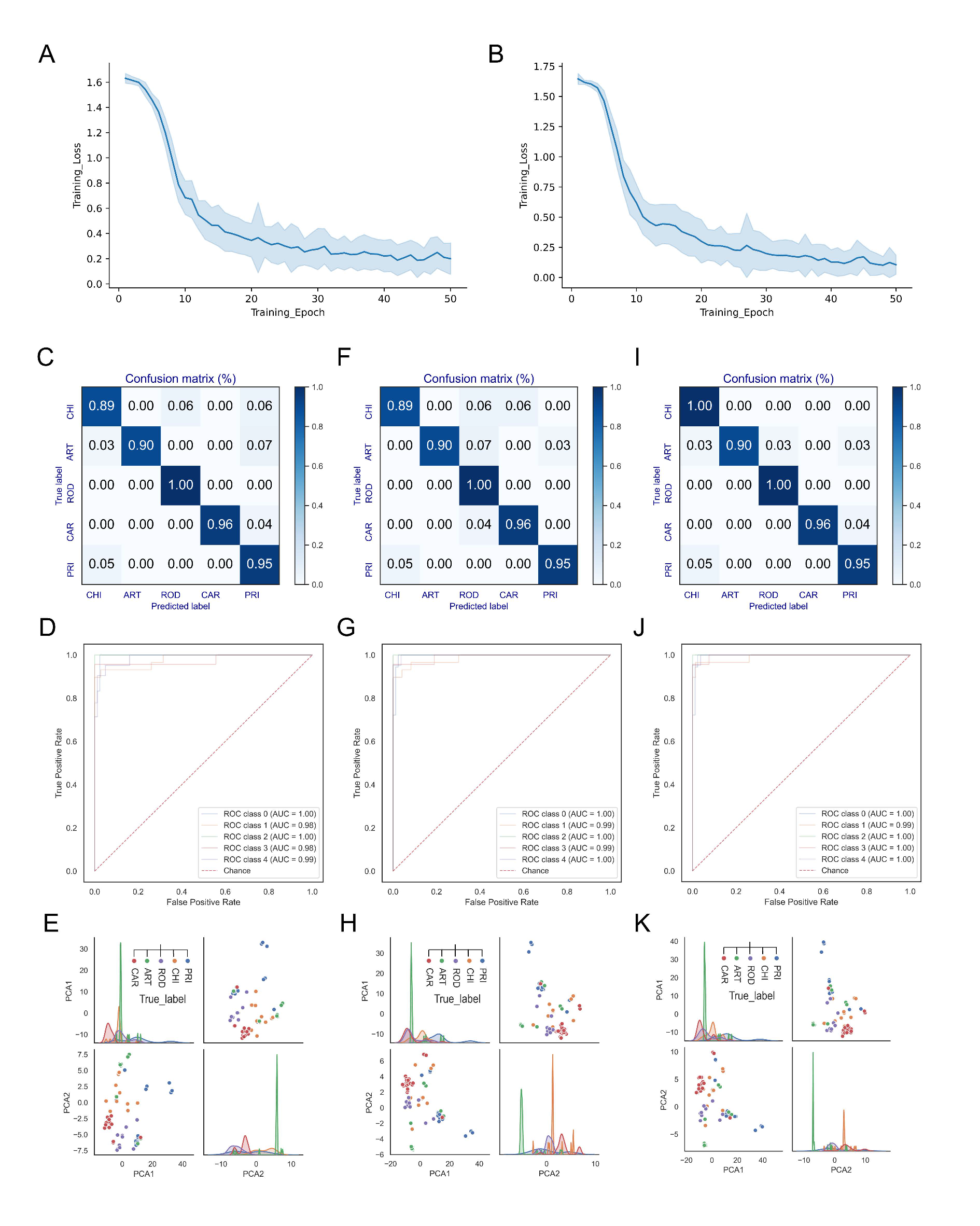
2.3. Establishment of Convolutional Neural Network (CNN) Models of ORF1ab and Spike Sequences
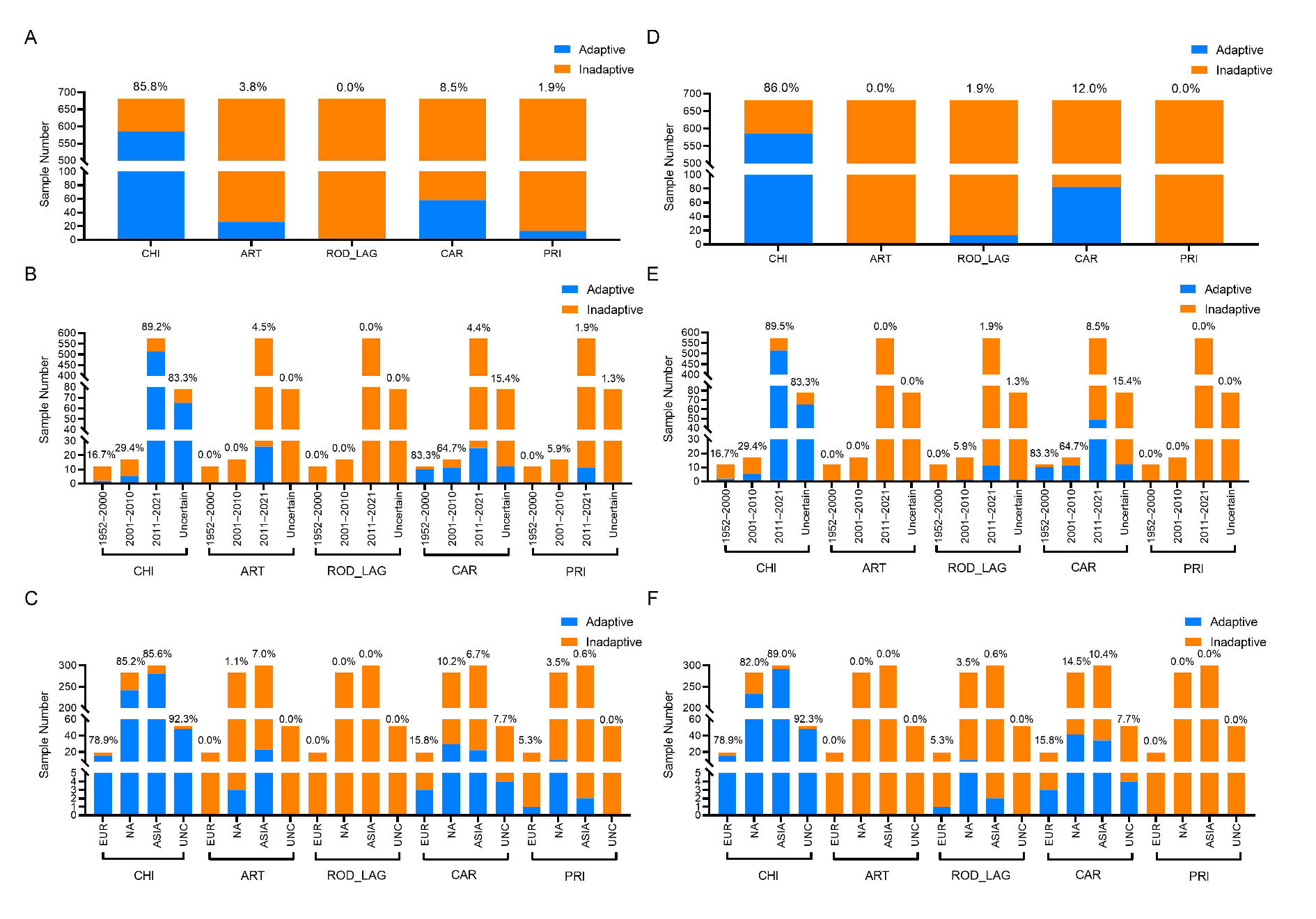
2.4. Prediction of the Adaptation Ratio of Suiformes CoVs to Various Hosts and the Spatiotemporal Distribution of the Adaptation Ratio
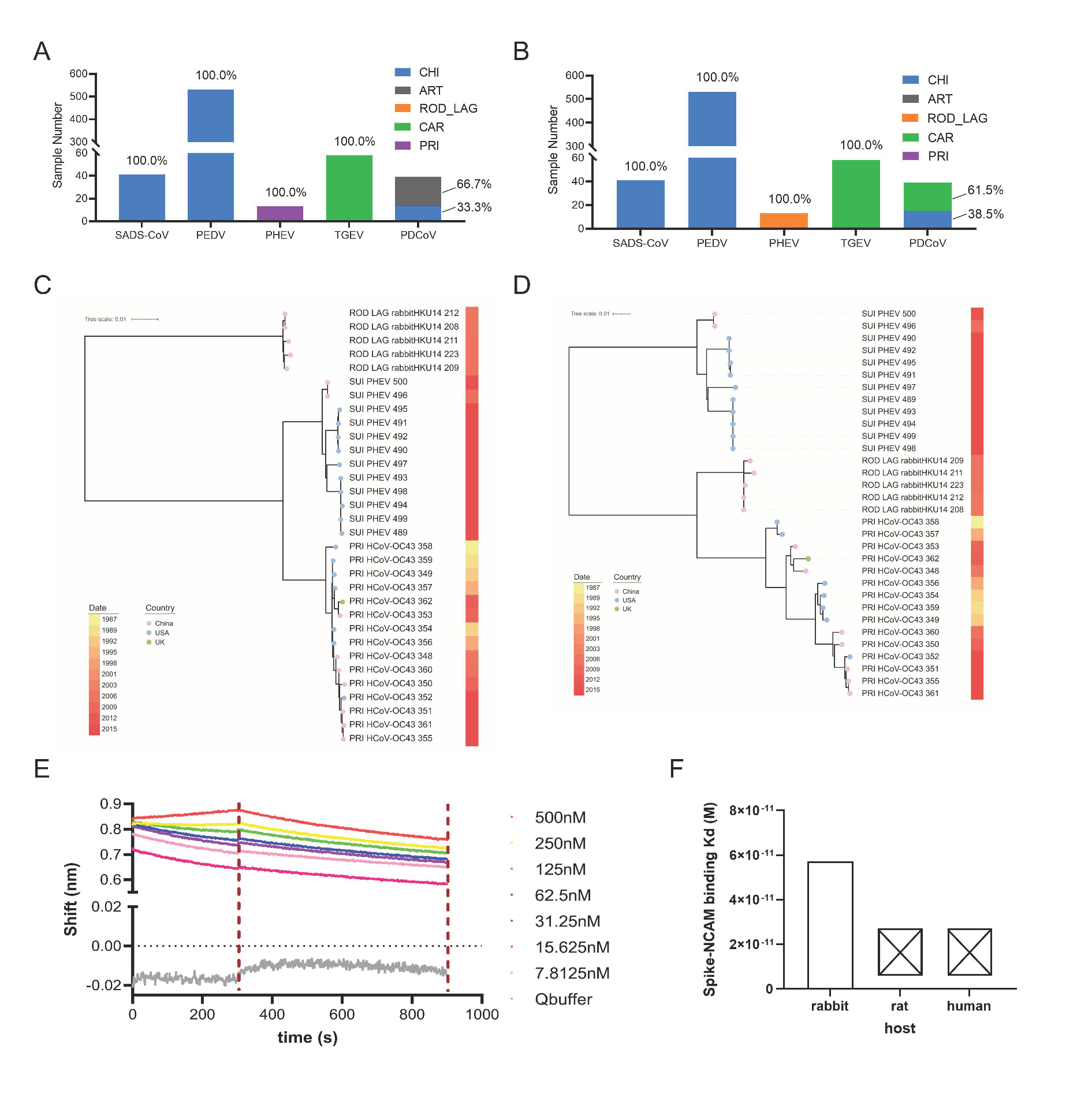
2.5. Phylogenetic Analysis of ORF1ab and Spike Genes
2.6. Biolayer Interferometry (BLI) Assay for RBD of PHE-CoV Spike and NCAM Interaction
3. Results
3.1. Prediction Pipeline of Adaptive Hosts of SUI CoVs
3.2. Unsupervised Learning of SUI and Other CoVs
3.3. Classification Effect of the DCR-Based CNN Models of ORF1ab and Spike
3.4. Adaptation Prediction of Suiformes CoVs Based on the CNN Models
3.5. Adaptation Prediction, Phylogenetic Analysis, and Receptor Binding Verification of PHEV
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Luk, H.K.H.; Wong, A.C.P.; Li, K.S.M.; Zhu, L.; He, Z.; Fung, J.; Chan, T.T.Y.; Fung, K.S.C.; Woo, P.C.Y. Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1542–1547. [Google Scholar] [CrossRef] [PubMed]
- Kobasa, D.; Jones, S.M.; Shinya, K.; Kash, J.C.; Copps, J.; Ebihara, H.; Hatta, Y.; Kim, J.H.; Halfmann, P.; Hatta, M.; et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 2007, 445, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef]
- Sanjuan, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef]
- Li, J.; Liu, B.; Chang, G.; Hu, Y.; Zhan, D.; Xia, Y.; Li, Y.; Yang, Y.; Zhu, Q. Virulence of H5N1 virus in mice attenuates after in vitro serial passages. Virol. J. 2011, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Hu, Y.; Sun, W.; Zhang, S.; Li, Y.; Wu, X.; Yang, Y.; Zhu, Q.; Jiang, T.; Li, J.; et al. Mouse lung-adapted mutation of E190G in hemagglutinin from H5N1 influenza virus contributes to attenuation in mice. J. Med. Virol. 2015, 87, 1816–1822. [Google Scholar] [CrossRef]
- Arai, Y.; Kawashita, N.; Daidoji, T.; Ibrahim, M.S.; El-Gendy, E.M.; Takagi, T.; Takahashi, K.; Suzuki, Y.; Ikuta, K.; Nakaya, T.; et al. Novel Polymerase Gene Mutations for Human Adaptation in Clinical Isolates of Avian H5N1 Influenza Viruses. PLoS Pathog. 2016, 12, e1005583. [Google Scholar] [CrossRef]
- Deng, Y.; Li, C.; Han, J.; Wen, Y.; Wang, J.; Hong, W.; Li, X.; Liu, Z.; Ye, Q.; Li, J.; et al. Phylogenetic and genetic characterization of a 2017 clinical isolate of H7N9 virus in Guangzhou, China during the fifth epidemic wave. Sci. China Life Sci. 2017, 60, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Jiang, L.; Li, J.; Zhao, Q.; Wang, J.; He, X.; Huang, S.; Wang, Q.; Zhao, Y.; Wang, G.; et al. Low Polymerase Activity Attributed to PA Drives the Acquisition of the PB2 E627K Mutation of H7N9 Avian Influenza Virus in Mammals. mBio 2019, 10, e1005583. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Qi, J.; Xiao, H.; Bi, Y.; Zhang, W.; Xu, Y.; Wang, F.; Shi, Y.; Gao, G.F. Avian-to-Human Receptor-Binding Adaptation by Influenza A Virus Hemagglutinin H4. Cell Rep. 2017, 20, 1201–1214. [Google Scholar] [CrossRef] [PubMed]
- Everest, H.; Billington, E.; Daines, R.; Burman, A.; Iqbal, M. The Emergence and Zoonotic Transmission of H10Nx Avian Influenza Virus Infections. mBio 2021, 12, e178521. [Google Scholar] [CrossRef]
- Anderson, T.K.; Chang, J.; Arendsee, Z.W.; Venkatesh, D.; Souza, C.K.; Kimble, J.B.; Lewis, N.S.; Davis, C.T.; Vincent, A.L. Swine Influenza A Viruses and the Tangled Relationship with Humans. Cold Spring Harb. Perspect. Med. 2021, 11, a038737. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Viboud, C.; Vincent, A.L.; Culhane, M.R.; Detmer, S.E.; Wentworth, D.E.; Rambaut, A.; Suchard, M.A.; Holmes, E.C.; Lemey, P. Global migration of influenza A viruses in swine. Nat. Commun. 2015, 6, 6696. [Google Scholar] [CrossRef]
- Ozawa, M.; Kawaoka, Y. Cross talk between animal and human influenza viruses. Annu. Rev. Anim. Biosci. 2013, 1, 21–42. [Google Scholar] [CrossRef]
- Yu, X.; Tsibane, T.; McGraw, P.A.; House, F.S.; Keefer, C.J.; Hicar, M.D.; Tumpey, T.M.; Pappas, C.; Perrone, L.A.; Martinez, O.; et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 2008, 455, 532–536. [Google Scholar] [CrossRef]
- Hilleman, M.R. Serologic responses to split and whole swine influenza virus vaccines in light of the next influenza pandemic. J. Infect. Dis. 1977, 136, S683–S685. [Google Scholar] [CrossRef]
- Li, J.; Tian, F.; Zhang, S.; Liu, S.S.; Kang, X.P.; Li, Y.D.; Wei, J.Q.; Lin, W.; Lei, Z.; Feng, Y.; et al. Genomic representation predicts an asymptotic host adaptation of bat coronaviruses using deep learning. Front. Microbiol. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Qin, P.; Wang, B.; Liu, Y.; Xu, G.H.; Peng, L.; Zhou, J.; Zhu, S.J.; Huang, Y.W. Broad Cross-Species Infection of Cultured Cells by Bat HKU2-Related Swine Acute Diarrhea Syndrome Coronavirus and Identification of Its Replication in Murine Dendritic Cells In Vivo Highlight Its Potential for Diverse Interspecies Transmission. J. Virol. 2019, 93, e01448-19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Li, J.; Zhou, Q.; Xu, Z.; Chen, L.; Zhang, Y.; Xue, C.; Wen, Z.; Cao, Y. A New Bat-HKU2-like Coronavirus in Swine, China, 2017. Emerg. Infect. Dis. 2017, 23, 1607–1609. [Google Scholar] [CrossRef] [PubMed]
- He, W.T.; Bollen, N.; Xu, Y.; Zhao, J.; Dellicour, S.; Yan, Z.; Gong, W.; Zhang, C.; Zhang, L.; Lu, M.; et al. Phylogeography Reveals Association between Swine Trade and the Spread of Porcine Epidemic Diarrhea Virus in China and across the World. Mol. Biol. Evol. 2022, 39, msab364. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.E.; Yount, B.L.; Graham, R.L.; Leist, S.R.; Hou, Y.J.; Dinnon, K.H., 3rd; Sims, A.C.; Swanstrom, J.; Gully, K.; Scobey, T.D.; et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA 2020, 117, 26915–26925. [Google Scholar] [CrossRef]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonnym, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Ye, X.; Chen, Y.; Zhu, X.; Guo, J.; Xie, D.; Hou, Z.; Xu, S.; Zhou, J.; Fang, L.; Wang, D.; et al. Cross-species transmission of deltacoronavirus and the origin of porcine deltacoronavirus. Evol. Appl. 2020, 13, 2246–2253. [Google Scholar] [CrossRef]
- Sit, T.H.C.; Brackman, C.J.; Ip, S.M.; Tam, K.W.S.; Law, P.Y.T.; To, E.M.W.; Yu, V.Y.T.; Sims, L.D.; Tsang, D.N.C.; Chu, D.K.W.; et al. Infection of dogs with SARS-CoV-2. Nature 2020, 586, 776–778. [Google Scholar] [CrossRef]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Moes, E.; Thoelen, I.; Wollants, E.; Lemey, P.; Vandamme, A.M.; Van Ranst, M. Complete genomic sequence of human coronavirus OC43: Molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 2005, 79, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv Virus Res 2018, 100, 163–188. [Google Scholar] [CrossRef]
- Kiyuka, P.K.; Agoti, C.N.; Munywoki, P.K.; Njeru, R.; Bett, A.; Otieno, J.R.; Otieno, G.P.; Kamau, E.; Clark, T.G.; Van Der Hoek, L.; et al. Human Coronavirus NL63 Molecular Epidemiology and Evolutionary Patterns in Rural Coastal Kenya. J. Infect. Dis. 2018, 217, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, J.G.; Vennema, H.; Godeke, G.J.; Horzinek, M.C.; Rottier, P.J. Nucleotide sequence and expression of the spike (S) gene of canine coronavirus and comparison with the S proteins of feline and porcine coronaviruses. J. Gen. Virol. 1994, 75, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Du, Q.; Feng, C.; Ma, L.; Zhang, Z. CompoDynamics: A comprehensive database for characterizing sequence composition dynamics. Nucleic Acids Res. 2021, 50, D962–D969. [Google Scholar] [CrossRef]
- Forsberg, R.; Christiansen, F.B. A codon-based model of host-specific selection in parasites, with an application to the influenza A virus. Mol Biol Evol. 2003, 20, 1252–1259. [Google Scholar] [CrossRef]
- Charles, H.; Calevro, F.; Vinuelas, J.; Fayard, J.M.; Rahbe, Y. Codon usage bias and tRNA over-expression in Buchnera aphidicola after aromatic amino acid nutritional stress on its host Acyrthosiphon pisum. Nucleic Acids Res. 2006, 34, 4583–4592. [Google Scholar] [CrossRef]
- Babayan, S.A.; Orton, R.J.; Streicker, D.G. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 2018, 362, 577–580. [Google Scholar] [CrossRef]
- Chen, F.; Wu, P.; Deng, S.; Zhang, H.; Hou, Y.; Hu, Z.; Zhang, J.; Chen, X.; Yang, J.R. Dissimilation of synonymous codon usage bias in virus-host coevolution due to translational selection. Nat. Ecol. Evol. 2020, 4, 589–600. [Google Scholar] [CrossRef]
- Hausser, J.; Mayo, A.; Keren, L.; Alon, U. Central dogma rates and the trade-off between precision and economy in gene expression. Nat. Commun. 2019, 10, 68. [Google Scholar] [CrossRef]
- Chen, J.; Gao, K.; Wang, R.; Wei, G.W. Prediction and mitigation of mutation threats to COVID-19 vaccines and antibody therapies. Chem Sci. 2021, 12, 6929–6948. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, S.; Li, B.; Hu, Y.; Kang, X.P.; Wu, X.Y.; Huang, M.T.; Li, Y.C.; Zhao, Z.P.; Qin, C.F.; et al. Machine Learning Methods for Predicting Human-Adaptive Influenza A Viruses Based on Viral Nucleotide Compositions. Mol. Biol. Evol. 2020, 37, 1224–1236. [Google Scholar] [CrossRef] [PubMed]
- Contu, L.; Balistreri, G.; Domanski, M.; Uldry, A.C.; Mühlemann, O. Characterisation of the Semliki Forest Virus-host cell interactome reveals the viral capsid protein as an inhibitor of nonsense-mediated mRNA decay. PLoS Pathog. 2021, 17, e1009603. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Annu Rev Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Duret, L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000, 16, 287–289. [Google Scholar] [CrossRef]
- Alley, E.C.; Khimulya, G.; Biswas, S.; AlQuraishi, M.; Church, G.M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 2019, 16, 1315–1322. [Google Scholar] [CrossRef]
- Hie, B.; Zhong, E.D.; Berger, B.; Bryson, B. Learning the language of viral evolution and escape. Science 2021, 371, 284–288. [Google Scholar] [CrossRef]
- Rao, R.; Bhattacharya, N.; Thomas, N.; Duan, Y.; Chen, X.; Canny, J.; Abbeel, P.; Song, Y.S. Evaluating Protein Transfer Learning with TAPE. Adv. Neural. Inf Process Syst. 2019, 32, 9689–9701. [Google Scholar]
- Xia, X. Extreme Genomic CpG Deficiency in SARS-CoV-2 and Evasion of Host Antiviral Defense. Mol. Biol. Evol. 2020, 37, 2699–2705. [Google Scholar] [CrossRef]
- Zahradník, J.; Marciano, S.; Shemesh, M.; Zoler, E.; Harari, D.; Chiaravalli, J.; Meyer, B.; Rudich, Y.; Li, C.; Marton, I.; et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol. 2021, 6, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Pucci, F.; Rooman, M. Prediction and Evolution of the Molecular Fitness of SARS-CoV-2 Variants: Introducing SpikePro. Viruses 2021, 13, 935. [Google Scholar] [CrossRef]
- Li, J.; Wu, Y.N.; Zhang, S.; Kang, X.P.; Jiang, T. Deep learning based on biologically interpretable genome representation predicts two types of human adaptation of SARS-CoV-2 variants. Brief. Bioinform. 2022, 23, bbac036. [Google Scholar] [CrossRef]
- Nan, B.G.; Zhang, S.; Li, Y.C.; Kang, X.P.; Chen, Y.H.; Li, L.; Jiang, T.; Li, J. Convolutional Neural Networks Based on Sequential Spike Predict the High Human Adaptation of SARS-CoV-2 Omicron Variants. Viruses 2022, 14, 1072. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data-from vision to reality. Euro. Surveill. 2017, 22, 30494. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242-5. [Google Scholar] [CrossRef]
- Dong, B.; Gao, W.; Lu, H.; Zhao, K.; Ding, N.; Liu, W.; Zhao, J.; Lan, Y.; Tang, B.; Jin, Z.; et al. A small region of porcine hemagglutinating encephalomyelitis virus spike protein interacts with the neural cell adhesion molecule. Intervirology 2015, 58, 130–137. [Google Scholar] [CrossRef]
- Gao, W.; He, W.; Zhao, K.; Lu, H.; Ren, W.; Du, C.; Chen, K.; Lan, Y.; Song, D.; Gao, F. Identification of NCAM that interacts with the PHE-CoV spike protein. Virol. J. 2010, 7, 254. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Xiao, Y.; Liu, J.; Wang, D.; Li, F.; Wang, C.; Li, C.; Zhu, J.; Song, J.; Sun, H.; et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. USA 2020, 117, 17204–17210. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef]
- Jung, K.; Hu, H.; Saif, L.J. Porcine deltacoronavirus infection: Etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016, 226, 50–59. [Google Scholar] [CrossRef]
- Brierley, L.; Fowler, A. Predicting the animal hosts of coronaviruses from compositional biases of spike protein and whole genome sequences through machine learning. PLoS Pathog. 2021, 17, e1009149. [Google Scholar] [CrossRef] [PubMed]
- Si, F.; Jiang, L.; Yu, R.; Wei, W.; Li, Z. Study on the Characteristic Codon Usage Pattern in Porcine Epidemic Diarrhea Virus Genomes and Its Host Adaptation Phenotype. Front. Microbiol. 2021, 12, 738082. [Google Scholar] [CrossRef]
- Bahir, I.; Fromer, M.; Prat, Y.; Linial, M. Viral adaptation to host: A proteome-based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 2009, 5, 311. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Zhang, S.; Kang, X.; Feng, Y.; Li, Y.; Nie, M.; Li, Y.; Chen, Y.; Zhao, S.; Jiang, T.; et al. Risk Assessment of the Possible Intermediate Host Role of Pigs for Coronaviruses with a Deep Learning Predictor. Viruses 2023, 15, 1556. https://doi.org/10.3390/v15071556
Jiang S, Zhang S, Kang X, Feng Y, Li Y, Nie M, Li Y, Chen Y, Zhao S, Jiang T, et al. Risk Assessment of the Possible Intermediate Host Role of Pigs for Coronaviruses with a Deep Learning Predictor. Viruses. 2023; 15(7):1556. https://doi.org/10.3390/v15071556
Chicago/Turabian StyleJiang, Shuyang, Sen Zhang, Xiaoping Kang, Ye Feng, Yadan Li, Maoshun Nie, Yuchang Li, Yuehong Chen, Shishun Zhao, Tao Jiang, and et al. 2023. "Risk Assessment of the Possible Intermediate Host Role of Pigs for Coronaviruses with a Deep Learning Predictor" Viruses 15, no. 7: 1556. https://doi.org/10.3390/v15071556
APA StyleJiang, S., Zhang, S., Kang, X., Feng, Y., Li, Y., Nie, M., Li, Y., Chen, Y., Zhao, S., Jiang, T., & Li, J. (2023). Risk Assessment of the Possible Intermediate Host Role of Pigs for Coronaviruses with a Deep Learning Predictor. Viruses, 15(7), 1556. https://doi.org/10.3390/v15071556






