-
 Molecular Detection of Various Non-Seasonal, Zoonotic Influenza Viruses Using BioFire FilmArray and GenXpert Diagnostic Platforms
Molecular Detection of Various Non-Seasonal, Zoonotic Influenza Viruses Using BioFire FilmArray and GenXpert Diagnostic Platforms -
 Primary HSV-2 Infection in an Immunocompromised Patient Reveals High Diversity of Drug-Resistance Mutations in the Viral DNA Polymerase
Primary HSV-2 Infection in an Immunocompromised Patient Reveals High Diversity of Drug-Resistance Mutations in the Viral DNA Polymerase -
 Spatiotemporal Characterization of Changes in the Respiratory Tract and the Nervous System, Including the Eyes in SARS-CoV-2-Infected K18-hACE2 Mice
Spatiotemporal Characterization of Changes in the Respiratory Tract and the Nervous System, Including the Eyes in SARS-CoV-2-Infected K18-hACE2 Mice -
 The Role of Prion Protein in Reelin/Dab1 Signaling: Implications for Neurodegeneration
The Role of Prion Protein in Reelin/Dab1 Signaling: Implications for Neurodegeneration -
 A Fluorescent Reporter Virus Toolkit for Interrogating Enterovirus Biology and Host Interactions
A Fluorescent Reporter Virus Toolkit for Interrogating Enterovirus Biology and Host Interactions
Journal Description
Viruses
Viruses
is a peer-reviewed, open access journal of virology, published monthly online by MDPI. The Spanish Society for Virology (SEV), Canadian Society for Virology (CSV), Italian Society for Virology (SIV-ISV), Australasian Virology Society (AVS), Brazilian Society for Virology (BSV) and others are affiliated with Viruses and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Virology) / CiteScore - Q1 (Virology/Infectious Diseases)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 18.6 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Companion journal: Zoonotic Diseases.
Impact Factor:
3.5 (2024);
5-Year Impact Factor:
3.7 (2024)
Latest Articles
Discovery of Landscape Phage Probes Against Cellular Communication Network Factor 1 (CCN1/Cyr61)
Viruses 2025, 17(9), 1273; https://doi.org/10.3390/v17091273 - 19 Sep 2025
Abstract
Detection of cancer biomarkers at the earliest stages of disease progression is commonly assumed to extend the overall quality of life for cancer patients as the result of earlier clinical management of the disease. Therefore, there is an urgent need for the development
[...] Read more.
Detection of cancer biomarkers at the earliest stages of disease progression is commonly assumed to extend the overall quality of life for cancer patients as the result of earlier clinical management of the disease. Therefore, there is an urgent need for the development of standardized, sensitive, robust, and commonly available screening and diagnostic tools for detecting the earliest signals of neoplastic pathology progression. Recently, a new paradigm of cancer control, known as multi-cancer detection (MCD), evolved, which measures the composition of cancer-related molecular analytes in the patient’s fluids using minimally invasive techniques. In this respect, the “Holy Grail” of cancer researchers and bioengineers for decades has been composing a repertoire or molecular sensing probes that would allow for the diagnosis, prognosis, and monitoring of cancer diseases via their interaction with cell-secreted and cell-associated cancer antigens and biomarkers. Therefore, the current trend in screening and detection of cancer-related pathologies is the development of portable biosensors for mobile laboratories and individual use. Phage display, since its conception by George Smith 40 years ago, has emerged as a premier tool for molecular evolution in molecular biology with widespread applications including identification and screening of cancer biomarkers, such as Circulating Cellular Communication Network Factor 1 (CCN1), an extracellular matrix-associated signaling protein responsible for a variety of cellular functions and has been shown to be overexpressed as part of the response to various pathologies including cancer. We hypothesize that CCN1 protein can be used as a soluble marker for the early detection of breast cancer in a multi-cancer detection (MCD) platform. However, validated probes have not been identified to date. Here, we screened the multi-billion clone landscape phage display library for phages interacting specifically with immobilized CCN1 protein. Through our study, we discovered a panel of 26 different phage-fused peptides interacting selectively with CCN1 protein that can serve for development of a novel phage-based diagnostic platform to monitor changes in CCN1 serum concentration by liquid biopsy.
Full article
(This article belongs to the Special Issue Phage Display in Cancer Diagnosis and Screening)
Open AccessArticle
Deep Learning-Based Automatic Segmentation and Analysis of Mitochondrial Damage by Zika Virus and SARS-CoV-2
by
Brianda Alexia Agundis-Tinajero, Miguel Ángel Coronado-Ipiña, Ignacio Lara-Hernández, Rodrigo Aparicio-Antonio, Anita Aguirre-Barbosa, Gisela Barrera-Badillo, Nidia Aréchiga-Ceballos, Irma López-Martínez, Claudia G Castillo, Vanessa Labrada-Martagón, Mauricio Comas-García and Aldo Rodrigo Mejía-Rodríguez
Viruses 2025, 17(9), 1272; https://doi.org/10.3390/v17091272 - 19 Sep 2025
Abstract
Viruses can induce various mitochondrial morphological changes, which are associated with the type of immune response. Therefore, characterization and analysis of mitochondrial ultrastructural changes could provide insights into the kind of immune response elicited, especially when compared to uninfected cells. However, this analysis
[...] Read more.
Viruses can induce various mitochondrial morphological changes, which are associated with the type of immune response. Therefore, characterization and analysis of mitochondrial ultrastructural changes could provide insights into the kind of immune response elicited, especially when compared to uninfected cells. However, this analysis is highly time-consuming and susceptible to observer bias. This work presents the development of a deep learning-based approach for the automatic identification, segmentation, and analysis of mitochondria from thin-section transmission electron microscopy images of cells infected with two SARS-CoV-2 variants or the Zika virus, utilizing a convolutional neural network with a U-Net architecture. A comparison between manual and automatic segmentations, along with morphological metrics, was performed, yielding an accuracy greater than 85% with no statistically significant differences between the manual and automatic metrics. This approach significantly reduces processing time and enables a prediction of the immune response to viral infections by allowing the detection of both intact and damaged mitochondria. Therefore, the proposed deep learning-based tool may represent a significant advancement in the study and understanding of cellular responses to emerging pathogens. Additionally, its applicability could be extended to the analysis of other organelles, thereby opening up new opportunities for automated studies in cell biology.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
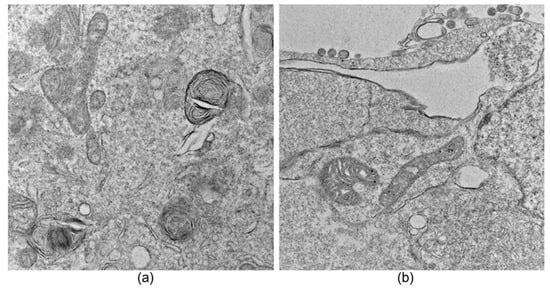
Figure 1
Open AccessArticle
Linking Pollution and Viral Risk: Detection of Dioxins and Coronaviruses in Cats and Dogs
by
Francesco Serra, Silvia Canzanella, Sergio Brandi, Gerardo Picazio, Anna Maria Pugliese, Luca Del Sorbo, Gianluca Miletti, Enza Ragosta, Emanuela Sannino, Filomena Fiorito, Mauro Esposito, Esterina De Carlo, Giovanna Fusco and Maria Grazia Amoroso
Viruses 2025, 17(9), 1271; https://doi.org/10.3390/v17091271 - 19 Sep 2025
Abstract
Viral and chemical analyses were performed on 80 dead cats and 51 dead dogs from the Campania Region (Southern Italy), with the aim of evaluating in vivo the potential correlation between coronavirus (CoV) infections and levels of environmental pollutants such as dioxins and
[...] Read more.
Viral and chemical analyses were performed on 80 dead cats and 51 dead dogs from the Campania Region (Southern Italy), with the aim of evaluating in vivo the potential correlation between coronavirus (CoV) infections and levels of environmental pollutants such as dioxins and PCSs (PCDD/F, DL-PCB and NDL-PCB). The overall viral prevalence was 16.3% in cats and 23.5% in dogs. Both feline coronavirus (FCoV) and canine coronavirus (CCoV) were identified, with variable detection rates in all the other organs investigated, supporting studies that provide evidence of systemic viral spread. The highest prevalence of coronaviruses (CoVs) was observed in Naples (19.2% for FCoV; 30.7% for CCoV) and Caserta (11.1% for FCoV; 50.0% for CCoV), areas that include municipalities with the highest Municipality Index of Environmental Pressure (MIEP) scores. Chemical analyses showed that DL-PCBs were present at more elevated concentrations in CoV-infected dogs and cats than in non-infected animals, whereas ∑NDL-PCB and ∑PCDD/F were detected in greater amounts in non-infected subjects. Among PCDDs, the congener 2,3,7,8-TCDD displayed different distribution patterns between infected and non-infected animals. In cats, 70.0% of FCoV-positive individuals had 2,3,7,8-TCDD levels above the limit of quantification (LOQ), compared with 38.0% of FCoV-negative cats. In dogs, 78.0% of CCoV-infected animals exceeded the LOQ, compared with 20.0% of non-infected ones; this difference was statistically significant. The results of the study suggest that elevated levels of 2,3,7,8-TCDD may be associated with CCoV infection and replication in dogs, suggesting a possible relationship between environmental pollution and susceptibility to coronavirus infections.
Full article
(This article belongs to the Section Animal Viruses)
Open AccessArticle
Isolation of Porcine Adenovirus Serotype 5 and Construction of Recombinant Virus as a Vector Platform for Vaccine Development
by
Qianhua He, Jun Wu, Zhilong Bian, Yuan Sun and Jingyun Ma
Viruses 2025, 17(9), 1270; https://doi.org/10.3390/v17091270 - 19 Sep 2025
Abstract
Porcine adenovirus serotype 5 (PAdV-5) is an emerging viral vector platform for veterinary vaccines; however, its genomic plasticity and essential replication elements remain incompletely characterized. This study reports the isolation and reverse genetic manipulation of a novel PAdV-5 strain (GD84) from diarrheic piglets
[...] Read more.
Porcine adenovirus serotype 5 (PAdV-5) is an emerging viral vector platform for veterinary vaccines; however, its genomic plasticity and essential replication elements remain incompletely characterized. This study reports the isolation and reverse genetic manipulation of a novel PAdV-5 strain (GD84) from diarrheic piglets in China. PCR screening of 167 clinical samples revealed a PAdV-5 detection rate of 38.3% (64/167), with successful isolation on ST cells after three blind passages. The complete GD84 genome is 32,620 bp in length and exhibited 99.0% nucleotide identity to the contemporary strain Ino5, but only 97.0% to the prototype HNF-70. It features an atypical GC content of 51.0% and divergent structural genes—most notably the hexon gene (89% identity to HNF-70)—suggesting altered immunogenicity. Using Red/ET recombineering, we established a rapid (less than 3 weeks) reverse genetics platform and generated four E3-modified recombinants: ΔE3-All-eGFP, ΔE3-12.5K-eGFP, ΔE3-12.5K+ORF4-eGFP, and E3-Insert-eGFP. Crucially, the ΔE3-All-eGFP construct (complete E3 deletion) failed to be rescued, while constructs preserving the 12.5K open reading frame (ORF) yielded replication-competent viruses with sustained eGFP expression over three serial passages and titers over 107.0 TCID50/mL. Fluorescence intensity was inversely correlated with genome size, as the full-length E3-Insert-eGFP virus showed reduced expression compared with the ΔE3 variants. Our work identifies the 12.5K ORF as essential for PAdV-5 replication and provides an optimized vaccine engineering platform that balances genomic payload capacity with replicative fitness.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
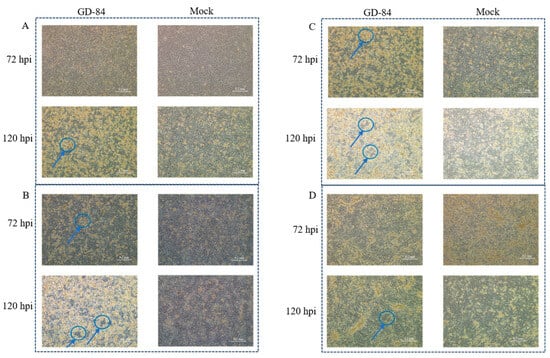
Figure 1
Open AccessReview
Insect-Specific Viruses and Their Emerging Role in Plant Disease Mitigation
by
Jianing Lei, Jingna Yuan, Mengnan Chen and Qianzhuo Mao
Viruses 2025, 17(9), 1269; https://doi.org/10.3390/v17091269 - 19 Sep 2025
Abstract
Insect vectors play a pivotal role in the emergence and dissemination of plant viral diseases. Beyond their function in transmitting plant viruses, these insects harbor diverse insect-specific viruses (ISVs). Advances in high-throughput sequencing (HTS) have uncovered virus diversity and prevalence in insects that
[...] Read more.
Insect vectors play a pivotal role in the emergence and dissemination of plant viral diseases. Beyond their function in transmitting plant viruses, these insects harbor diverse insect-specific viruses (ISVs). Advances in high-throughput sequencing (HTS) have uncovered virus diversity and prevalence in insects that far exceed previous estimations. However, current knowledge of ISVs remains predominantly limited to genomic sequencing information. Investigating the fundamental biology of ISVs, their effects on insect physiology, and their modulation of vector competence is critical for deciphering complex virus–virus and virus–insect interactions. Such research holds substantial promise for developing innovative biocontrol strategies against plant viral pathogens. This review synthesizes current insights into the interplay between plant viruses and their insect vectors, explores the discovery and functional roles of ISVs, and discusses the potential application of ISVs in mitigating plant viral diseases. Understanding these dynamic relationships offers new avenues for sustainable plant disease management.
Full article
(This article belongs to the Section Viruses of Plants, Fungi and Protozoa)
►▼
Show Figures
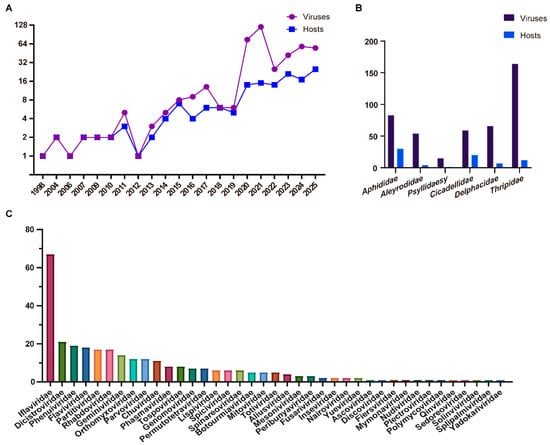
Figure 1
Open AccessReview
Tomato Bushy Stunt Virus (TBSV): From a Plant Pathogen to a Multifunctional Biotechnology Platform
by
Almas Madirov, Nurgul Iksat and Zhaksylyk Masalimov
Viruses 2025, 17(9), 1268; https://doi.org/10.3390/v17091268 - 19 Sep 2025
Abstract
Plant viruses have evolved from being viewed exclusively as pathogens into versatile and powerful tools for modern biotechnology. Among them, Tomato bushy stunt virus (TBSV) holds a special place due to its well-studied molecular biology and unique structural properties. This review systematizes the
[...] Read more.
Plant viruses have evolved from being viewed exclusively as pathogens into versatile and powerful tools for modern biotechnology. Among them, Tomato bushy stunt virus (TBSV) holds a special place due to its well-studied molecular biology and unique structural properties. This review systematizes the knowledge on TBSV’s dual role as a multifunctional platform. On one hand, we cover its application as a viral vector for the highly efficient expression of recombinant proteins in plants, as well as a tool for functional genomics, including Virus-Induced Gene Silencing (VIGS) and the delivery of CRISPR/Cas9 gene-editing components. On the other hand, we provide a detailed analysis of the use of the stable and monodisperse TBSV virion in nanobiotechnology. Its capsid serves as an ideal scaffold for creating next-generation vaccine candidates, platforms for targeted drug delivery to tumor cells, and as a building block for the programmable self-assembly of complex nanoarchitectures. In conclusion, key challenges limiting the widespread adoption of the platform are discussed, including the genetic instability of vectors and difficulties in scalable purification, along with promising strategies to overcome them.
Full article
(This article belongs to the Special Issue Application of Plant Viruses in Biotechnology)
►▼
Show Figures
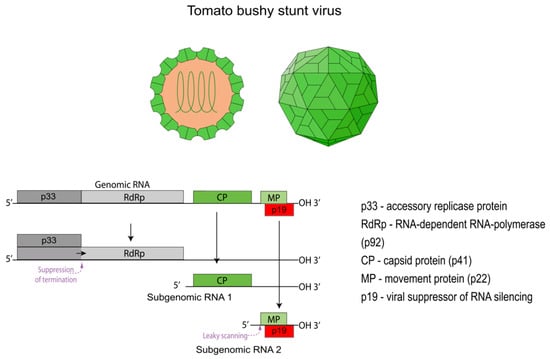
Figure 1
Open AccessArticle
Probing Viral Dark Matter: Comparative Genomics of Atypical Bacillus Phage YungSlug
by
Allison A. Johnson, Andrew Hale, Amine Sehnouni, Zainab Gbadamosi and Bret M. Boyd
Viruses 2025, 17(9), 1267; https://doi.org/10.3390/v17091267 - 19 Sep 2025
Abstract
Bacillus phage YungSlug is a novel phage with a genome that has limited homology to known viruses. To better understand this unique phage, we searched for close relatives of YungSlug using traditional comparative genomics approaches and a broad search of a large protein
[...] Read more.
Bacillus phage YungSlug is a novel phage with a genome that has limited homology to known viruses. To better understand this unique phage, we searched for close relatives of YungSlug using traditional comparative genomics approaches and a broad search of a large protein database. YungSlug shares only 9% genome alignment with Bacillus phage Nachito, its closest relative, and less than 1% with others in its subfamily. A search for homologs in the NCBI nr database was dominated by low percent identity homologs from viral, bacteria, and environmental sources, returning matches for only 50% of the predicted proteins in YungSlug’s genome. Additionally, a set of 21 conserved proteins was identified that may define a core gene set for the Spounavirinae subfamily of Herelleviridae. These findings highlight the diversity of phages infecting Bacillus and highlight gaps in our knowledge of the Bacillus-infecting phage community.
Full article
(This article belongs to the Special Issue Bacteriophage Diversity, 2nd Edition)
►▼
Show Figures
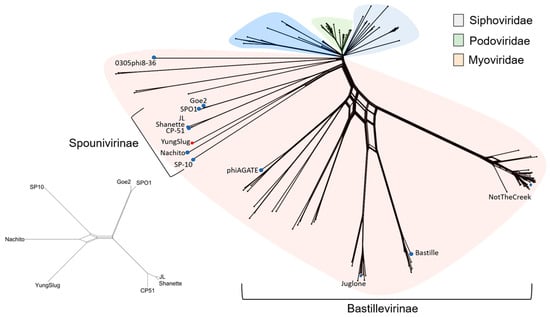
Figure 1
Open AccessArticle
Co-Detection of ADV, Influenza B, and HPIV: Independent Risk Factors for SMPP with Changes in NPIs
by
Linlin Huang and Ting Shi
Viruses 2025, 17(9), 1266; https://doi.org/10.3390/v17091266 - 19 Sep 2025
Abstract
Background: This study investigated the epidemiology of Mycoplasma pneumoniae (MP) in children with acute respiratory tract infections (ARTIs) and explored the risk factors for severe mycoplasma pneumoniae pneumonia (SMPP) in children. Methods: A retrospective analysis was conducted on 36,380 children with acute respiratory
[...] Read more.
Background: This study investigated the epidemiology of Mycoplasma pneumoniae (MP) in children with acute respiratory tract infections (ARTIs) and explored the risk factors for severe mycoplasma pneumoniae pneumonia (SMPP) in children. Methods: A retrospective analysis was conducted on 36,380 children with acute respiratory infections who underwent multiplex real-time polymerase chain reaction (RT-PCR) assays for nine respiratory pathogens from September 2021 to November 2024. Results: A total of 36,380 children with ARTIs were enrolled in this study. The co-detection rate of MP with other pathogens was significantly higher in the post-NPIs period than in the NPIs period (36.5% vs. 25.7%, p < 0.01). Multivariate regression identified the detection of influenza A virus (InfA), InfB, human parainfluenza virus (HPIV), human bocaparvovirus (HBoV), human rhinovirus (HRV), adenovirus (ADV), human respiratory syncytial virus (HRSV), and human metapneumovirus (HMPV) as protective factors against MP epidemics (p < 0.01); meanwhile, older age, the cancellation of NPIs, and summer–autumn seasons were found to be risk factors. After adjusting for sex, age, period, season, and pathogens, InfB (OR: 3.009, 95%CI: 1.041–8.697, p = 0.042), HPIV (OR: 2.226, 95%CI: 1.170–4.235, p = 0.015), and ADV (OR: 2.035, 95%CI: 1.105–3.750, p = 0.023) were identified as independent risk factors for SMPP. Conclusions: These findings highlight post-NPI shifts in MP epidemiology and identify ADV, InfB, and HPIV as early warning markers for SMPP.
Full article
(This article belongs to the Special Issue Respiratory Viral Pathogenesis and Host-Microbe Crosstalk: From Bench to Bedside)
►▼
Show Figures
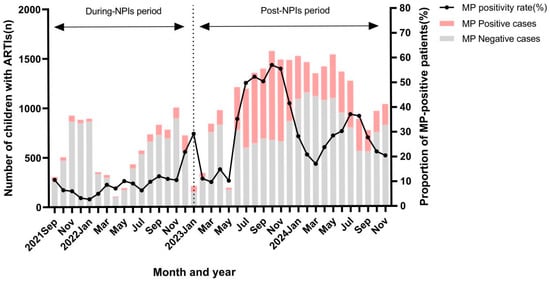
Figure 1
Open AccessArticle
Prevalence and Genetic Diversity of Torque teno felis virus (FcTTV) in Domestic Cats from Kazakhstan
by
Gulzhan Yessembekova, Bolat Abdigulov, Alexandr Shevtsov, Asylulan Amirgazin, Sarsenbay Abdrakhmanov, Elena Shevtsova, Symbat Bolysbekkyzy, Salima Baduanova and Alexandr Shustov
Viruses 2025, 17(9), 1265; https://doi.org/10.3390/v17091265 - 19 Sep 2025
Abstract
Anelloviruses have a broad mammalian host range, including Torque teno felis virus (FcTTV), a felid-associated member that remains undercharacterized. This is the first comprehensive study of FcTTV in domestic cats in Central Asia. We analyzed blood samples from 206 domestic cats from the
[...] Read more.
Anelloviruses have a broad mammalian host range, including Torque teno felis virus (FcTTV), a felid-associated member that remains undercharacterized. This is the first comprehensive study of FcTTV in domestic cats in Central Asia. We analyzed blood samples from 206 domestic cats from the large city of Astana, Kazakhstan, collected in 2023–2024. Using nested PCR we identified 63 FcTTV-positive samples (30.6% prevalence), and the sequences were compared to global reference strains. Potential demographic associations (sex and age) were assessed. The study revealed an overall FcTTV prevalence of 30.6%. Infection rates showed no significant sex-related differences: ages varied 4–168 months. ORF1 sequencing revealed multiple FcTTV variants in 27% of samples, with no demographic links. Phylogenetic analysis revealed distinct patterns at both nucleotide and amino acid levels: 3 groups of nucleotide sequences (max divergence 21.68%; intra-cluster 5.15–6.8%), and 3 clusters of amino acid sequences (max divergence 16.81%; intra-cluster 2.82–6.68%). Deletions were found in ORF1 in some variants. Global phylogeny aligned clusters with Asian/European strains (90–98% identity), confirming FcTTV1 affiliation and inter-regional transmission. Our study of FcTTV in Kazakhstan reveals moderate virus prevalence with considerable genetic diversity across viral strains and frequent co-infections with multiple variants.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures

Figure 1
Open AccessArticle
Rapid Visual Detection of Senecavirus A Based on RPA-CRISPR/Cas12a System with Canonical or Suboptimal PAM
by
Xinrui Zhao, Genghong Jiang, Qinyi Ruan, Yunjie Qu, Xiaoyu Yang, Yongyan Shi, Dedong Wang, Jianwei Zhou, Jue Liu and Lei Hou
Viruses 2025, 17(9), 1264; https://doi.org/10.3390/v17091264 - 18 Sep 2025
Abstract
Senecavirus A (SVA) is an emerging pathogen responsible for vesicular lesions and neonatal mortality in swine. In the absence of effective vaccines or therapeutics, early and accurate diagnosis is essential for controlling SVA outbreaks. Although nucleic acid-based detection methods are commonly employed, there
[...] Read more.
Senecavirus A (SVA) is an emerging pathogen responsible for vesicular lesions and neonatal mortality in swine. In the absence of effective vaccines or therapeutics, early and accurate diagnosis is essential for controlling SVA outbreaks. Although nucleic acid-based detection methods are commonly employed, there remains a pressing need for rapid, convenient, highly sensitive, and specific diagnostic tools. Here, we developed a two-pot assay combining recombinase polymerase amplification (RPA) with CRISPR/Cas12a containing crRNA targeting canonical protospacer adjacent motifs (PAMs) for simple, rapid, and visual identification of SVA in clinical samples. Subsequently, we successfully streamlined this system into a one-pot assay by selecting a specially designed crRNA targeting suboptimal PAM and integrating RPA amplification reagents and CRISPR/Cas12a detection components into a single reaction system in one tube. The developed methods exhibited diagnostic specificity, showing no cross-reactivity with four major swine viruses, while showing remarkable sensitivity with a lower detection limit of just two copies. Clinical validation in field samples using these two methods revealed perfect agreement (100% concordance) with conventional quantitative PCR (qPCR) results (sample size, n = 28), with both assays completing detection within 30 min. These results demonstrate that both the one-pot and two-pot RPA-CRISPR/Cas12a assays offer a reliable and efficient method for detecting SVA in this pilot study. Despite the limited sample size, the assays combine rapid reaction time with high sensitivity and specificity, showing great potential for future diagnostic applications.
Full article
(This article belongs to the Section General Virology)
►▼
Show Figures
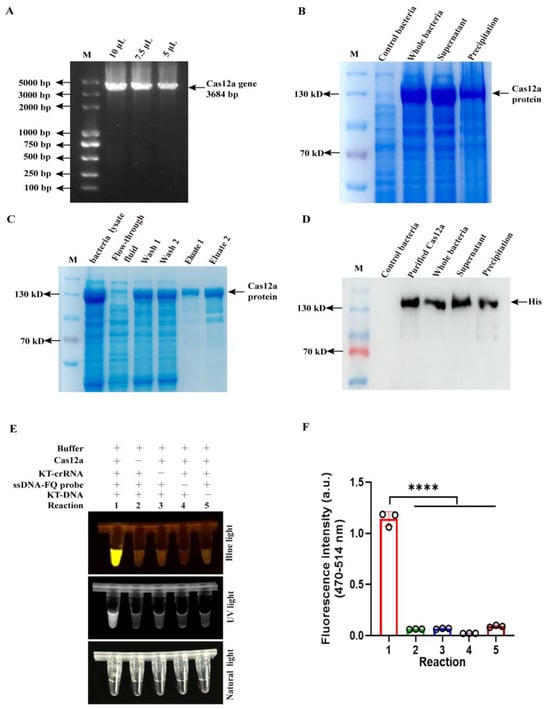
Figure 1
Open AccessArticle
Neutral Impact of SARS-CoV-2 Coinfection on the Recombination-Driven Evolution of Endemic HCoV-OC43
by
Xueling Zheng, Yinyan Zhou, Yue Yu, Shi Cheng, Feifei Cao, Zhou Sun, Jun Li and Xinfen Yu
Viruses 2025, 17(9), 1263; https://doi.org/10.3390/v17091263 - 18 Sep 2025
Abstract
Knowledge gaps exist on whether SARS-CoV-2 co-infection alters recombination frequency or induces phylogenetic incongruities in endemic β-coronaviruses (HCoV-OC43, HCoV-HKU1), limiting our understanding of cross-species evolution. Among 7213 COVID-19 and 1590 non-COVID-19 acute respiratory cases (2021–2022) screened via multiplex PCR, β-coronavirus co-infections (SARS-CoV-2 +
[...] Read more.
Knowledge gaps exist on whether SARS-CoV-2 co-infection alters recombination frequency or induces phylogenetic incongruities in endemic β-coronaviruses (HCoV-OC43, HCoV-HKU1), limiting our understanding of cross-species evolution. Among 7213 COVID-19 and 1590 non-COVID-19 acute respiratory cases (2021–2022) screened via multiplex PCR, β-coronavirus co-infections (SARS-CoV-2 + HCoV-OC43/HKU1) and single HCoV-OC43/HKU1 infections were identified. Whole-genome sequencing (Illumina NovaSeq) was performed. Phylogenies were reconstructed using Bayesian inference (MrBayes). Recombination was assessed via Bootscan analysis (SimPlot). Co-infection prevalence was low (0.51%, mainly HCoV-HKU1: 0.28%, HCoV-OC43: 0.11%). HCoV-OC43 diverged into lineage 1 (genotype K) and a novel recombinant lineage 2 (genotypes F/J/G/I segments), exhibiting accelerated evolution. HCoV-HKU1 remained genetically stable (genotype B). Co-infection status did not influence evolutionary outcomes. While SARS-CoV-2 co-infection may favor transmission of endemic HCoVs, their evolution appears driven by population-level selection, not co-infection. HCoV-OC43 underwent recombination-driven diversification, contrasting sharply with HCoV-HKU1’s stasis, highlighting distinct evolutionary strategies. Integrated genomic and clinical surveillance is critical for tracking coronavirus adaptation.
Full article
(This article belongs to the Special Issue COVID-19 Complications and Co-infections)
►▼
Show Figures
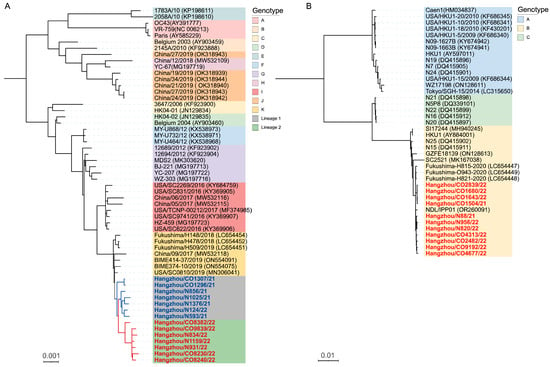
Figure 1
Open AccessReview
Porcine Parvovirus in China: Recent Advances, Epidemiology, and Vaccine Strategies
by
Yunchao Liu, Yumei Chen, Yanli Shang, Xiuli Deng and Huifang Hao
Viruses 2025, 17(9), 1262; https://doi.org/10.3390/v17091262 - 18 Sep 2025
Abstract
Porcine parvovirus (PPV), a non-envelope single-stranded DNA virus, causes severe reproductive disorders in swine worldwide, characterized by fetal mortality, mummification, and reduced boar fertility. As a highly prevalent pathogen in Chinese swine herds, PPV imposes substantial economic burdens on intensive pig production systems.
[...] Read more.
Porcine parvovirus (PPV), a non-envelope single-stranded DNA virus, causes severe reproductive disorders in swine worldwide, characterized by fetal mortality, mummification, and reduced boar fertility. As a highly prevalent pathogen in Chinese swine herds, PPV imposes substantial economic burdens on intensive pig production systems. This review systematically synthesizes recent advances in PPV virology, focusing on genomic evolution of emerging strains (PPV1–PPV8), epidemiological dynamics of emerging strains, molecular pathogenesis, and novel diagnostic tools. Furthermore, we critically evaluate current vaccine strategies, highlighting their limitations in cross-protective efficacy and viral shedding control. By integrating multi-omics insights with immunological profiling, this work delineates actionable pathways for next-generation vaccine design and proposes a roadmap for rational antigen selection. This review consolidates foundational knowledge and establishes a translational bridge between basic virology and prevention and control of porcine parvovirus, addressing critical gaps in porcine reproductive disease management.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
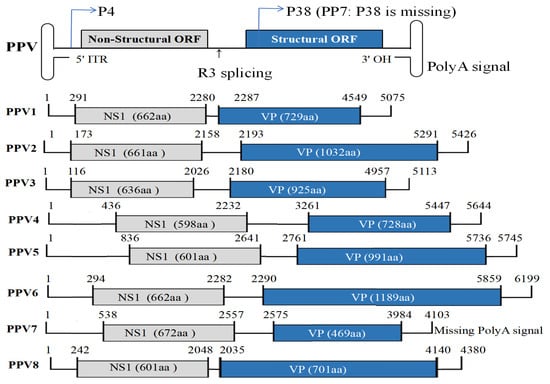
Figure 1
Open AccessArticle
Predicted Structures of Ceduovirus Adhesion Devices Highlight Unique Architectures Reminiscent of Bacterial Secretion System VI
by
Adeline Goulet, Jennifer Mahony, Douwe van Sinderen and Christian Cambillau
Viruses 2025, 17(9), 1261; https://doi.org/10.3390/v17091261 - 18 Sep 2025
Abstract
Bacteriophages, or phages, are sophisticated nanomachines that efficiently infect bacteria. Their infection of lactic acid bacteria (LAB) used in fermentation can lead to significant industrial losses. Among phages that infect monoderm bacteria, those with siphovirion morphology characterized by a long, non-contractile tail are
[...] Read more.
Bacteriophages, or phages, are sophisticated nanomachines that efficiently infect bacteria. Their infection of lactic acid bacteria (LAB) used in fermentation can lead to significant industrial losses. Among phages that infect monoderm bacteria, those with siphovirion morphology characterized by a long, non-contractile tail are predominant. The initial stage of phage infection involves precise host recognition and binding. To achieve this, phages feature host adhesion devices (HADs) located at the distal end of their tails, which have evolved to recognize specific proteinaceous or saccharidic receptors on the host cell wall. Ceduovirus represents a group of unique lytic siphophages that specifically infect the LAB Lactococcus lactis by targeting proteinaceous receptors. Despite having compact genomes, most of their structural genes are poorly annotated and the architecture and function of their HADs remain unknown. Here we used AlphaFold3 to explore the Ceduovirus HADs and their interaction with the host. We show that Ceduovirus HADs exhibit unprecedented features among bacteriophages infecting Gram+, share structural similarities with bacterial secretion system VI, and combine both saccharide and protein-binding modules. Moreover, we could annotate the majority of Ceduovirus genes encoding structural proteins by leveraging their predicted structures, highlighting AlphaFold’s significant contribution to phage genome annotation.
Full article
(This article belongs to the Section Bacterial Viruses)
►▼
Show Figures
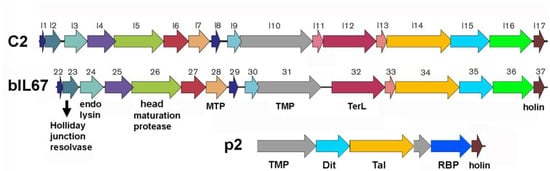
Figure 1
Open AccessArticle
Structural and Functional Characterization of Porcine Adeno-Associated Viruses
by
Austin Nelson, Mario Mietzsch, Jane Hsi, Julia Eby, Paul Chipman and Robert McKenna
Viruses 2025, 17(9), 1260; https://doi.org/10.3390/v17091260 - 18 Sep 2025
Abstract
Current gene therapy treatments utilizing adeno-associated virus (AAV) vectors are based on capsids of primate origin. However, pre-existing neutralizing anti-AAV antibodies, that are present in a significant portion of the population, can lead to vector inactivation and reduced therapeutic efficacy. Advances in DNA
[...] Read more.
Current gene therapy treatments utilizing adeno-associated virus (AAV) vectors are based on capsids of primate origin. However, pre-existing neutralizing anti-AAV antibodies, that are present in a significant portion of the population, can lead to vector inactivation and reduced therapeutic efficacy. Advances in DNA sequencing have facilitated the discovery of many AAVs from non-primate species, including isolates from pigs, which exhibit up to 50% capsid protein sequence divergence, compared to primate AAV serotypes. In this study, AAVs isolated from porcine tissues (AAVpo.1 and AAVpo.6) were selected for structural characterization due to their low capsid protein VP1 sequence identity compared to each other and to AAV9. The AAV vectors were produced via the standard triple transfection system in HEK293 cells using AAV2 rep to package AAV2-ITR vector genomes and were purified by iodixanol density gradient ultracentrifugation. The capsid structures of AAVpo.1 and AAVpo.6 were determined using cryo-electron microscopy and then compared to each other in addition to the AAV5 and AAV9 structures. Given that porcine AAVpo.6 has been reported to infect human cells and the ability to cross the blood–brain barrier, the functional characterization was focused on the identification of a potential glycan receptor utilized by the porcine capsids. Additionally, the porcine AAV capsid reactivity to human derived anti-AAV antibodies was assessed to evaluate the potential for these capsids to be used as alternative vectors for gene therapy, particularly for patients with pre-existing immunity to primate-derived AAV serotypes.
Full article
(This article belongs to the Special Issue Porcine Viruses 2025)
►▼
Show Figures
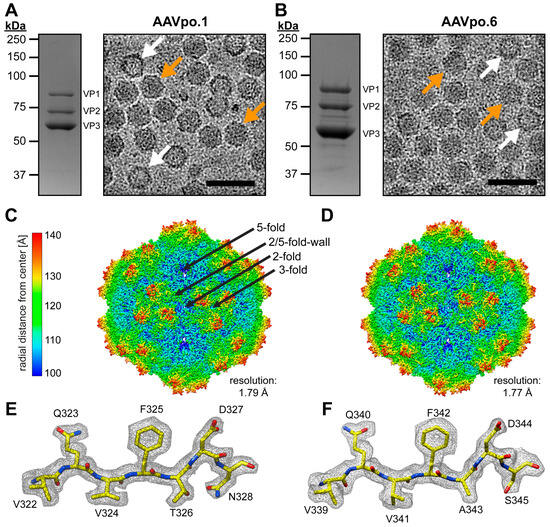
Figure 1
Open AccessArticle
Comparative Performance of Digital PCR and Real-Time RT-PCR in Respiratory Virus Diagnostics
by
Irene Bianconi, Giovanna Viviana Pellecchia, Elisabetta Maria Incrocci, Fabio Vittadello, Maira Nicoletti and Elisabetta Pagani
Viruses 2025, 17(9), 1259; https://doi.org/10.3390/v17091259 - 18 Sep 2025
Abstract
Background: Respiratory viral infections pose a major global health burden, and molecular diagnostics such as Real-Time RT-PCR have revealed frequent co-infections. However, precise quantification of viral RNA remains challenging. Digital PCR (dPCR) offers absolute quantification without standard curves and may improve diagnostic
[...] Read more.
Background: Respiratory viral infections pose a major global health burden, and molecular diagnostics such as Real-Time RT-PCR have revealed frequent co-infections. However, precise quantification of viral RNA remains challenging. Digital PCR (dPCR) offers absolute quantification without standard curves and may improve diagnostic accuracy. This study compares dPCR and Real-Time RT-PCR in detecting and quantifying influenza A, influenza B, respiratory syncytial virus (RSV), and SARS-CoV-2 during the 2023–2024 tripledemic. Methods: A total of 123 respiratory samples were analysed and stratified by cycle threshold (Ct) values into high, medium, and low viral load categories. Both dPCR and Real-Time RT-PCR were used to quantify and compare viral loads across these categories. Results: dPCR demonstrated superior accuracy, particularly for high viral loads of influenza A, influenza B, and SARS-CoV-2, and for medium loads of RSV. It showed greater consistency and precision than Real-Time RT-PCR, especially in quantifying intermediate viral levels. Conclusions: These findings highlight the potential of dPCR to enhance respiratory virus diagnostics and support a better understanding of co-infection dynamics. Nonetheless, its routine implementation is currently limited by higher costs and reduced automation compared to Real-Time RT-PCR.
Full article
(This article belongs to the Section General Virology)
►▼
Show Figures
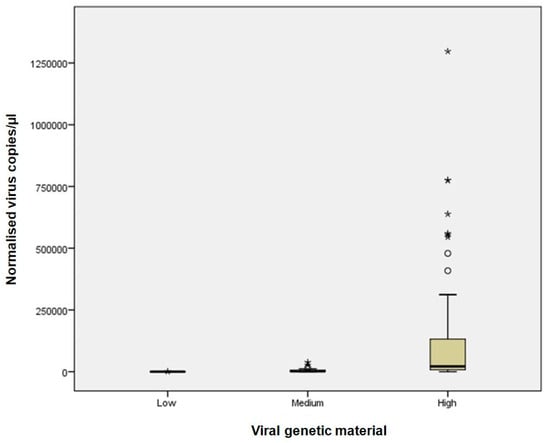
Figure 1
Open AccessArticle
Molecular Detection of Yellow Fever Virus in Haemagogus janthinomys Mosquitoes (Diptera: Culicidae) in a Rural Settlement in the State of Pará, Brazilian Amazon, 2024
by
Joaquim Pinto Nunes Neto, Daniel Damous Dias, Bruna Laís Sena do Nascimento, Sandro Patroca da Silva, Sâmia Luzia Sena da Silva, Lúcia Aline Moura Reis, Hanna Carolina Farias Reis, Fábio Silva da Silva, Lucas Henrique da Silva e Silva, Durval Bertram Rodrigues Vieira, Roberto Carlos Feitosa Brandão, Wallace Oliveira Rosário, Francisco Amilton dos Santos Paiva, José Wilson Rosa Júnior, Bruno Tardelli Diniz Nunes, Lívia Carício Martins, Lívia Medeiros Neves Casseb and Ana Cecília Ribeiro Cruz
Viruses 2025, 17(9), 1258; https://doi.org/10.3390/v17091258 - 18 Sep 2025
Abstract
Yellow fever (YF) is an acute and potentially fatal hemorrhagic disease caused by the Yellow Fever virus (YFV), endemic to sub-Saharan Africa and several tropical countries, including Brazil. In Brazil, the Amazon region is considered the main endemic area. YFV is maintained in
[...] Read more.
Yellow fever (YF) is an acute and potentially fatal hemorrhagic disease caused by the Yellow Fever virus (YFV), endemic to sub-Saharan Africa and several tropical countries, including Brazil. In Brazil, the Amazon region is considered the main endemic area. YFV is maintained in a sylvatic cycle involving Neotropical primates and mosquitoes of the genera Haemagogus and Sabethes, acting as primary and secondary vectors, respectively. In March 2024, entomovirological surveillance was conducted in Santa Bárbara do Pará, Pará, Brazil. A total of 286 mosquitoes were collected, classified into 13 species across nine genera, and grouped into 33 pools. Seventeen pools were tested by RT-qPCR for Orthoflavivirus (YFV, DENV, WNV, SLEV), Alphavirus (CHIKV, MAYV), and Orthobunyavirus (OROV). YFV was detected in four Haemagogus janthinomys pools, with Ct values ranging from 22.2 to 27.9. Metagenomic sequencing confirmed the presence of YFV with assigned reads and >99% protein identity. Notably, the detection occurred without human cases or primate deaths, enabling timely vaccination of the local population. These findings confirm YFV circulation in forested areas of the Belém metropolitan region and reaffirm Hg. janthinomys as a key vector. Our study reinforces the relevance of early entomovirological surveillance and preventive strategies, such as vaccination, to mitigate yellow fever reemergence.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
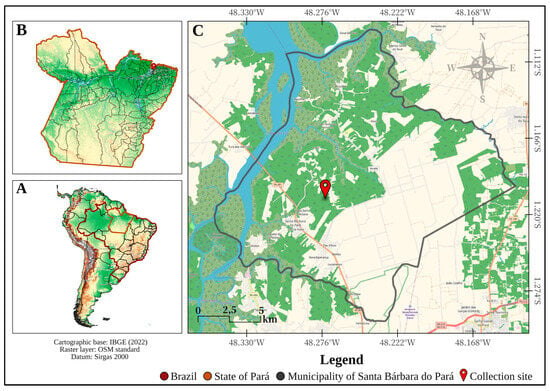
Figure 1
Open AccessCommentary
Mpox Epidemics: A Call to Restore Humanity’s Lost Herd Immunity to Orthopoxviruses
by
Misaki Wayengera, Henry Kyobe-Bosa, Winters Muttamba, Olushayo Oluseun Olu, Abdou Salam Gueye, Nicaise Ndembi, Neema Kamara, Morenike Oluwatoyin Folayan, Bruce Kirenga, Sitong Luo, Qingyu Li and Chikwe Ihekweazu
Viruses 2025, 17(9), 1257; https://doi.org/10.3390/v17091257 - 18 Sep 2025
Abstract
Global efforts to eradicate smallpox—an Orthopoxvirus infection—began in the mid-20th century, with the last naturally occurring case reported in 1977. This was achieved through global solidarity efforts that expanded the smallpox eradication vaccination program. Approximately 50 years following the cessation of mass smallpox
[...] Read more.
Global efforts to eradicate smallpox—an Orthopoxvirus infection—began in the mid-20th century, with the last naturally occurring case reported in 1977. This was achieved through global solidarity efforts that expanded the smallpox eradication vaccination program. Approximately 50 years following the cessation of mass smallpox vaccination and in the absence of access to a sustainable boosting program, the population immunologically naïve to Orthopoxviruses has increased significantly. With increasing global movements and travels, we argue that the emergence of two back-to-back yet distinct mpox epidemics in the 21st century is a sign of humanity’s lost herd immunity to Orthopoxviruses. This needs concerted efforts to restore.
Full article
(This article belongs to the Special Issue Mpox (Monkeypox): From Neglected Tropical Disease to Emerging Global Pathogen)
►▼
Show Figures

Figure 1
Open AccessReview
BK Polyomavirus-Associated Nephropathy and Hemorrhagic Cystitis in Transplant Recipients—What We Understand and What Remains Unclear
by
Tang-Her Jaing, Yi-Lun Wang and Tsung-Yen Chang
Viruses 2025, 17(9), 1256; https://doi.org/10.3390/v17091256 - 17 Sep 2025
Abstract
The reactivation of BK polyomavirus (BKPyV) during severe immunosuppression plays a crucial role in two significant syndromes observed in transplant recipients: BK polyomavirus-associated nephropathy (BKPyVAN) in kidney transplant patients and BK polyomavirus-associated hemorrhagic cystitis (BKPyV-HC) in hematopoietic cell transplant (HCT) recipients. This review
[...] Read more.
The reactivation of BK polyomavirus (BKPyV) during severe immunosuppression plays a crucial role in two significant syndromes observed in transplant recipients: BK polyomavirus-associated nephropathy (BKPyVAN) in kidney transplant patients and BK polyomavirus-associated hemorrhagic cystitis (BKPyV-HC) in hematopoietic cell transplant (HCT) recipients. This review aims to summarize the current understanding and lingering ambiguity by looking at three primary questions: (1) In cases with BKPyV-related illnesses in transplant patients, which diagnostic methods have the best track record of accuracy and success? (2) Which therapy approaches have the best track records of safety and efficacy in real-world clinical settings? (3) What can immunological research teach us about the development of future tailored treatments? Diagnosis involves the patient’s appearance, ruling out other potential causes, and employing quantitative PCR to identify active viral replication in urine or plasma. BKPyV-HC can vary from self-limited hematuria to potentially fatal bleeding, while BKPyVAN may lead to loss and dysfunction of the allograft. Reducing immunosuppression remains the key aspect of treatment. However, the effectiveness of antivirals (such cidofovir and leflunomide) is not always the same, and supporting measures depend on the syndrome. Researchers are looking into new immunotherapies, such as virus-specific cytotoxic T cells. Due to the intricate viro-immunopathology and lack of defined treatment regimens, future initiatives should focus on prospective studies to establish validated thresholds, enhance management algorithms, and integrate immune surveillance into individualized therapy.
Full article
(This article belongs to the Special Issue Viral Immunology in Transplant Patients)
►▼
Show Figures
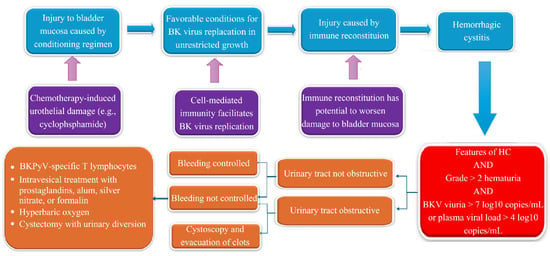
Figure 1
Open AccessReview
Hot and Cold HCC: Uncoupling Viral Oncogenesis and Therapy
by
Laura Sneller, Keshav Mathur, Shyam Kottilil and Poonam Mathur
Viruses 2025, 17(9), 1255; https://doi.org/10.3390/v17091255 - 17 Sep 2025
Abstract
Hepatocellular carcinoma (HCC) is rising in incidence globally. It is the sixth most common cancer and the third leading cause of cancer-related mortality worldwide. Infection with hepatitis B and/or C virus is a significant risk factor for developing HCC. These viruses exert their
[...] Read more.
Hepatocellular carcinoma (HCC) is rising in incidence globally. It is the sixth most common cancer and the third leading cause of cancer-related mortality worldwide. Infection with hepatitis B and/or C virus is a significant risk factor for developing HCC. These viruses exert their carcinogenicity in both direct and indirect ways, including induction of immune exhaustion with prolonged antigen exposure. Therefore, the best therapeutic option for HCC is prevention, i.e., Hepatitis B vaccination and treatment of viral hepatitis. However, when HCC develops because of viral hepatitis or other etiologies, long-lasting effects on the immune system remain even after viral suppression, which affect the response to HCC therapy. Recent studies have suggested a “hot” and “cold” model for HCC, in which the two kinds of HCC tumors have very distinct tumor microenvironments. The microenvironment for hot HCC makes these tumors amenable to immunotherapy with checkpoint inhibitors. Therefore, converting cold HCC tumors to hot tumors may make them susceptible to immunotherapy. In this review, we provide an overview of HCC epidemiology and prevention, an overview of tumor microenvironments of hot and cold HCC, the proposed mechanisms for converting cold tumors to hot tumors, and a concise summary of the evidence for combination checkpoint inhibitor therapy for HCC.
Full article
(This article belongs to the Special Issue Translational Research in Virology)
►▼
Show Figures
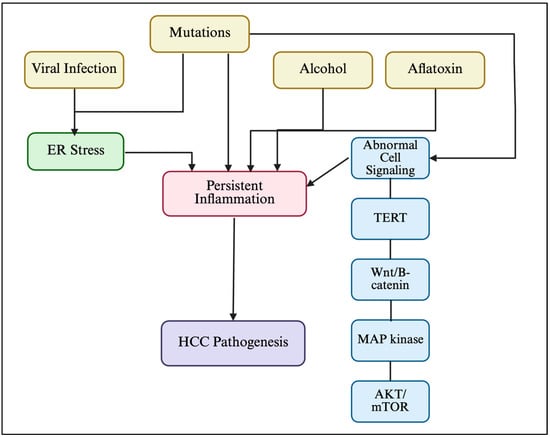
Figure 1
Open AccessCase Report
Real-World Experience with Long-Acting Injectable Cabotegravir/Rilpivirine in HIV Patients with Unsuppressed Viral Load
by
Marcello Trizzino, Luca Pipitò, Pierluigi Francesco Salvo, Federica Zimmerhofer, Andrea Cicero, Gianmaria Baldin, Claudia Conti, Claudia Gioè, Simona Di Giambenedetto and Antonio Cascio
Viruses 2025, 17(9), 1254; https://doi.org/10.3390/v17091254 - 17 Sep 2025
Abstract
Long-acting injectable cabotegravir/rilpivirine (CAB/RPV-LA) is currently approved as a maintenance therapy for people with HIV (PWH) who are virologically suppressed. However, growing real-world evidence highlights its potential role in more complex viremic populations traditionally considered ineligible. We present a case series of eight
[...] Read more.
Long-acting injectable cabotegravir/rilpivirine (CAB/RPV-LA) is currently approved as a maintenance therapy for people with HIV (PWH) who are virologically suppressed. However, growing real-world evidence highlights its potential role in more complex viremic populations traditionally considered ineligible. We present a case series of eight PWH treated at two tertiary centers in Italy, all of whom faced persistent viremia, adherence difficulties, malabsorption syndromes, or psychosocial barriers. Following the switch to CAB/RPV-LA, all patients, despite heterogeneous clinical profiles and baseline virological status, achieved and maintained virologic suppression, demonstrated improved adherence, and experienced no serious adverse events.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)

Journal Menu
► ▼ Journal Menu-
- Viruses Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
19 September 2025
MDPI Webinar | The Science Behind the Prize: 2025 Nobel Physiology or Medicine Roundtable, 6 October 2025
MDPI Webinar | The Science Behind the Prize: 2025 Nobel Physiology or Medicine Roundtable, 6 October 2025

10 September 2025
Meet Us at the 26th National Symposium on Environmental Microbiology, 26–29 September 2025, Shanghai, China
Meet Us at the 26th National Symposium on Environmental Microbiology, 26–29 September 2025, Shanghai, China

Topics

Conferences
Special Issues
Special Issue in
Viruses
Diversity and Evolution of Viruses in Ecosystem 2025
Guest Editor: René KalliesDeadline: 30 September 2025
Special Issue in
Viruses
JC Polyomavirus
Guest Editors: Sébastien Lhomme, Anne-Sophie L'HonneurDeadline: 30 September 2025
Special Issue in
Viruses
Molecular Epidemiology of SARS-CoV-2, 4th Edition
Guest Editor: Marta GiovanettiDeadline: 30 September 2025
Special Issue in
Viruses
Viral Strategies to Regulate Host Immunity or Signal Pathways 2nd Edition
Guest Editor: Bumsuk HahmDeadline: 30 September 2025
Topical Collections
Topical Collection in
Viruses
Mathematical Modeling of Viral Infection
Collection Editors: Amber M. Smith, Ruian Ke








