-
 Identification and Characterization of MmuPV1 Causing Papillomatosis Outbreak in an Animal Research Facility
Identification and Characterization of MmuPV1 Causing Papillomatosis Outbreak in an Animal Research Facility -
 Potential for Core Fucose-Targeted Therapy Against HBV Infection of Human Normal Hepatocytes
Potential for Core Fucose-Targeted Therapy Against HBV Infection of Human Normal Hepatocytes -
 Broadly Sarbecovirus-Neutralizing Antibodies Induced by Ancestral SARS-CoV-2 Infection
Broadly Sarbecovirus-Neutralizing Antibodies Induced by Ancestral SARS-CoV-2 Infection -
 Dysregulation of microRNAs in the Brains of Mice Infected with Powassan Virus
Dysregulation of microRNAs in the Brains of Mice Infected with Powassan Virus -
 Immune Responses and Replication of Rescued Torque Teno Virus (TTSuV1) in Mice
Immune Responses and Replication of Rescued Torque Teno Virus (TTSuV1) in Mice
Journal Description
Viruses
Viruses
is a peer-reviewed, open access journal of virology, published monthly online by MDPI. The Spanish Society for Virology (SEV), Canadian Society for Virology (CSV), Italian Society for Virology (SIV-ISV), Australasian Virology Society (AVS), Brazilian Society for Virology (BSV) and others are affiliated with Viruses and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Virology) / CiteScore - Q1 (Virology/Infectious Diseases)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 18.6 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Companion journal: Zoonotic Diseases.
Impact Factor:
3.5 (2024);
5-Year Impact Factor:
3.7 (2024)
Latest Articles
Special Issue: “Innate Immunity to Virus Infection, 2nd Edition”
Viruses 2025, 17(12), 1616; https://doi.org/10.3390/v17121616 (registering DOI) - 14 Dec 2025
Abstract
The frequent emergence of highly pathogenic viruses globally has persistently threatened global health [...]
Full article
(This article belongs to the Special Issue Innate Immunity to Virus Infection 2nd Edition)
Open AccessReview
CAR-T Cell Therapy for HIV Cure: Current Challenges, Advances and Future Directions
by
Monica-Daniela Padurariu-Covit, Costinela Georgescu, Mihaela Andreescu, Iulia Chiscop, Catalin Plesea-Condratovici and Manuela Arbune
Viruses 2025, 17(12), 1615; https://doi.org/10.3390/v17121615 (registering DOI) - 14 Dec 2025
Abstract
Antiretroviral therapy (ART) effectively suppresses HIV replication but fails to eradicate latent reservoirs, leading to viral rebound after interruption. Chimeric antigen receptor (CAR) T-cell therapy offers a potential strategy to achieve durable remission. A systematic PubMed search (July 2020–June 2025) identified 253 studies
[...] Read more.
Antiretroviral therapy (ART) effectively suppresses HIV replication but fails to eradicate latent reservoirs, leading to viral rebound after interruption. Chimeric antigen receptor (CAR) T-cell therapy offers a potential strategy to achieve durable remission. A systematic PubMed search (July 2020–June 2025) identified 253 studies on CAR-T therapy in HIV; 74 met inclusion criteria and were qualitatively analyzed. Preclinical data showed that CAR-T cells can recognize and eliminate infected cells, reach viral reservoirs, and persist long term, particularly when derived from hematopoietic stem cells. Dual-target and combination approaches with checkpoint inhibitors or latency-reversing agents enhanced antiviral efficacy. Early clinical studies confirmed safety and modest reservoir reduction. CAR-T cell therapy represents a promising step toward a functional HIV cure. Further optimization of design, integration with gene-editing technologies, and standardized clinical evaluation are required to confirm durable efficacy and safety.
Full article
(This article belongs to the Special Issue HIV Reservoirs, Latency, and the Factors Responsible)
►▼
Show Figures
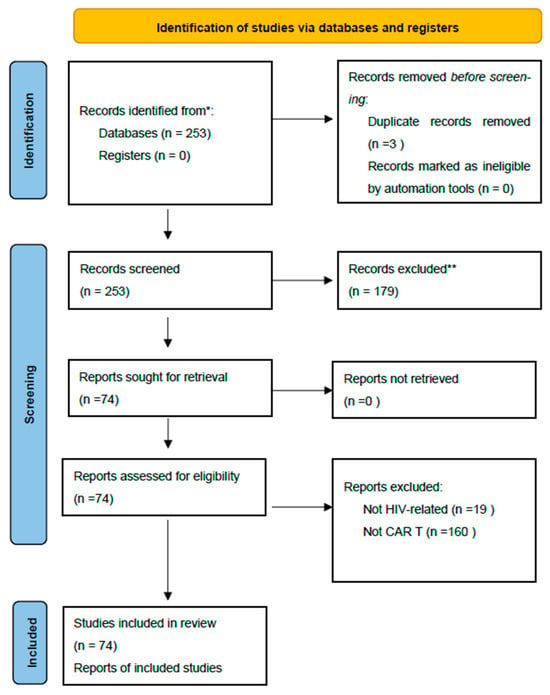
Figure 1
Open AccessArticle
Generating STEC-Specific Ackermannviridae Bacteriophages Through Tailspike Protein Chimerization
by
Jose Gil, John Paulson, Henriett Zahn, Matthew Brown, Minh M. Nguyen and Stephen Erickson
Viruses 2025, 17(12), 1614; https://doi.org/10.3390/v17121614 (registering DOI) - 14 Dec 2025
Abstract
Shiga toxin-producing Escherichia coli (STEC) pose a significant threat to public health and effective methods of detection are needed. The use of naturally occurring bacteriophages (phages) to detect E. coli has been well documented. However, detecting multiple serotypes at the same time often
[...] Read more.
Shiga toxin-producing Escherichia coli (STEC) pose a significant threat to public health and effective methods of detection are needed. The use of naturally occurring bacteriophages (phages) to detect E. coli has been well documented. However, detecting multiple serotypes at the same time often required multiple phages specific to individual serotypes. To limit the burden of complex cocktails, this study aimed to engineer phages with an expanded host range that allows each phage to contribute to detection across multiple STEC serogroups. Kutterviruses, in the Ackermannviridae family, contain four tailspike proteins (TSPs), each of which confers tropism to a different bacterial strain. The modular nature of TSPs allows for mixing receptor-binding domains from diverse phage types. The host range of the Kuttervirus CBA120 was modified by replacing its native tailspike proteins (TSPs) with chimeric versions incorporating receptor-binding domains from related and unrelated phages. A structure-guided approach was utilized to overcome minimal sequence similarity between donor and recipient phages and achieve novel functional TSP chimeras. Two engineered phage variants were created that collectively detect five STEC serogroups: O26, O45, O103, O111, and O157. Spotting and luciferase assays confirmed that the replacement TSPs were functional and the phages had acquired new host ranges. This study demonstrates the feasibility of engineering Ackermannviridae phages with customized host ranges for detecting multiple STEC strains. This approach has potential applications in developing improved phage-based bacterial detection, therapy, and biocontrol.
Full article
(This article belongs to the Special Issue Biotechnological Applications of Phage and Phage-Derived Proteins 2025)
►▼
Show Figures
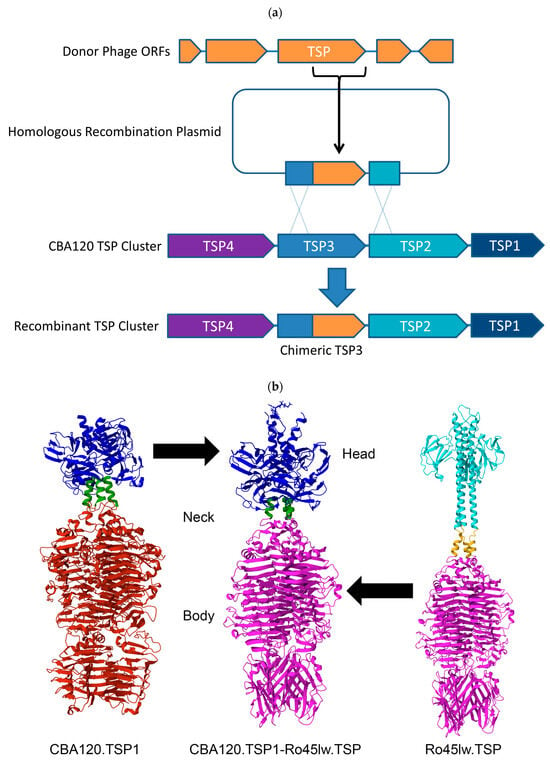
Figure 1
Open AccessArticle
Repurposing FDA-Approved Drugs as Hendra Virus RNA-Dependent RNA Polymerase Inhibitors: A Comprehensive Computational Drug Discovery Approach
by
Anjana C. Lalu, Varun Thachan Kundil, Bristow Ben Joseph, Radul R. Dev, Amritha Thaikkad, Suhail Subair, Rajesh Raju and Abhithaj Jayanandan
Viruses 2025, 17(12), 1613; https://doi.org/10.3390/v17121613 (registering DOI) - 13 Dec 2025
Abstract
Hendra virus (HeV) is a highly pathogenic zoonotic paramyxovirus that poses a serious threat to human and equine health, yet no approved antivirals or vaccines currently exist. RNA-dependent RNA polymerase (RdRp) of Hendra virus represents a critical and attractive target for antiviral drug
[...] Read more.
Hendra virus (HeV) is a highly pathogenic zoonotic paramyxovirus that poses a serious threat to human and equine health, yet no approved antivirals or vaccines currently exist. RNA-dependent RNA polymerase (RdRp) of Hendra virus represents a critical and attractive target for antiviral drug development, given its essential role in both viral genome replication and mRNA transcription. Due to the lack of a human homolog, it is more druggable and less likely to cause host toxicity. Its sequence conservation among related paramyxoviruses further highlights its potential for the development of broad-spectrum inhibitors. This study offers the first comprehensive computational analysis of the Hendra virus RdRp, potentially promising FDA-approved drugs as possible inhibitors. A homology model of RdRp was generated in the absence of experimental three-dimensional (3D) structure, followed by virtual screening and molecular dynamics (MD) simulations to evaluate the drug binding and stability. Based on the highest energy, four FDA-approved drugs selected were menadiol diphosphate (−49.88 kcal/mol), masoprocol (−39.69 kcal/mol), pamidronic acid (−34.29 kcal/mol), and dinoprostone (−46.90 kcal/mol). Furthermore, these compounds exhibited significant interactions with the catalytic GDNE motif. With strong conformational stability and pharmacokinetic profile, masoprocol and menadiol diphosphate showed the most stable and energetically favorable interactions within the RdRp active site. These findings suggest their potential as repurposed therapeutic candidates against Hendra virus infection and they provide a structural basis for the development of broad-spectrum paramyxovirus inhibitors, justifying additional experimental confirmation.
Full article
(This article belongs to the Special Issue Zoonotic and Vector-Borne Viral Diseases: 2nd Edition)
►▼
Show Figures
Figure 1
Open AccessOpinion
COVID-19 Double Annual Epidemic Peaks in Summer and in Winter from 2022, Irrespective of the Rate of Mask Wearing and Vaccination
by
Shinako Inaida, Richard E. Paul and Minsoo Kim
Viruses 2025, 17(12), 1612; https://doi.org/10.3390/v17121612 (registering DOI) - 13 Dec 2025
Abstract
Although vaccination for COVID-19 and mask wearing were two of the main preventive measures against infection, their impact is unclear. In the present study, by using national surveillance data in Japan, we compared the incidence rate and weekly case increase ratios of COVID-19
[...] Read more.
Although vaccination for COVID-19 and mask wearing were two of the main preventive measures against infection, their impact is unclear. In the present study, by using national surveillance data in Japan, we compared the incidence rate and weekly case increase ratios of COVID-19 with the domestic stocks of masks and vaccination coverage. The trajectory of epidemic growth increased rapidly in the summer of 2021, concomitant with the launch of the mass national vaccination program. The most rapid spread of the epidemic was found in 2022, approximately 6 months after the national mass vaccination started, with the emergence of the Omicron variant. From 2022, two annual epidemic peaks occurred with seasonal changes. Whilst the winter peak follows the expected seasonal trend in respiratory infections, the summer peak may reflect a combination of short-term herd immunity and behavioral patterns. Nevertheless, these epidemic peaks continued irrespective of vaccine coverage and mask use. Further analysis into the duration of protective efficacy of the vaccines and mask use is required.
Full article
(This article belongs to the Special Issue SARS-CoV-2, COVID-19 Pathologies, Long COVID, and Anti-COVID Vaccines)
Open AccessArticle
Molecular Testing in Organ Biopsies and Perfusion Fluid Samples from Severe Acute Respiratory Syndrome Coronavirus 2 Positive Donors
by
Evangelia Petrisli, Liliana Gabrielli, Carlo De Cillia, Andrea Liberatore, Giulia Piccirilli, Simona Venturoli, Alice Balboni, Eva Caterina Borgatti, Alessia Cantiani, Lamberto Manzoli, Nicola Alvaro and Tiziana Lazzarotto
Viruses 2025, 17(12), 1611; https://doi.org/10.3390/v17121611 (registering DOI) - 13 Dec 2025
Abstract
At the beginning of the COVID-19 pandemic, SARS-CoV-2-positive donors were not considered eligible for organ donation. The Italian National Transplant Centre has gradually introduced measures to prevent donor-to-recipient transmission of SARS-CoV-2 infection through organ transplantation. The current national screening protocol for deceased SARS-CoV-2-positive
[...] Read more.
At the beginning of the COVID-19 pandemic, SARS-CoV-2-positive donors were not considered eligible for organ donation. The Italian National Transplant Centre has gradually introduced measures to prevent donor-to-recipient transmission of SARS-CoV-2 infection through organ transplantation. The current national screening protocol for deceased SARS-CoV-2-positive donors recommends molecular testing of donor lower respiratory tract (LRT) samples, graft biopsies and organ perfusion fluids. The aim of the study is to describe the 3-year experience of protocol application in a northern region of Italy. From 1 January 2022 to 31 January 2025, a total of 132 samples were analyzed (29 liver biopsies, 35 kidney biopsies, 68 perfusion fluids) from 40 organ donors with an active or resolved SARS-CoV-2 infection. SARS-CoV-2 PCR on LRT samples was positive in 26/40 (65%) donors, negative in 11/40 (27.5%) cases and in the remaining 3 (7.5%) the PCR result was unknown. Overall, 65 organs were transplanted into 60 recipients. All processed graft biopsies and organ perfusion fluid samples tested negative for SARS-CoV-2 RNA. Our data suggest that the utilization of non-lung donors with resolved or active SARS-CoV-2 infections who died of other causes appears justified and safe.
Full article
(This article belongs to the Section Coronaviruses)
Open AccessArticle
Clinical and Immunological Recovery Trajectories in Severe COVID-19 Survivors: A 12-Month Prospective Follow-Up Study
by
Edita Strumiliene, Laura Malinauskiene, Jurgita Urboniene, Laimutė Jurgauskienė, Birutė Zablockienė and Ligita Jancoriene
Viruses 2025, 17(12), 1610; https://doi.org/10.3390/v17121610 - 12 Dec 2025
Abstract
Background: The link between clinical recovery and immune restoration after severe COVID-19 remains poorly defined. Although most survivors experience symptomatic improvement, persistent symptoms have been hypothesized to reflect ongoing immune dysregulation. Methods: This prospective cohort study followed 93 unvaccinated adults with RT-PCR-confirmed moderate-to-critical
[...] Read more.
Background: The link between clinical recovery and immune restoration after severe COVID-19 remains poorly defined. Although most survivors experience symptomatic improvement, persistent symptoms have been hypothesized to reflect ongoing immune dysregulation. Methods: This prospective cohort study followed 93 unvaccinated adults with RT-PCR-confirmed moderate-to-critical COVID-19 at 3, 6, and 12 months post-discharge. Clinical assessments used structured interviews to evaluate the persistent symptoms. Peripheral blood analyses were used to measure lymphocyte subsets, immunoglobulins, and complement components. Results: Clinical recovery was substantial; fatigue prevalence declined from 70.9% to 24.7% and dyspnea prevalence from 81.7% to 25.8% by 12 months (p < 0.001 for both). However, immune recovery exhibited divergent patterns. Activated T cells (CD3+HLA-DR+) decreased significantly (from 20% to 13%; p < 0.001), complement C3c levels paradoxically increased from 1.23 to 1.35 g/L (p < 0.001), and serum IgA increased by 32% (p = 0.003). NK cells remained stable overall but were persistently reduced in a subset (~25%) of patients, particularly among those with fatigue and dyspnea. Critical illness was associated with slower T-cell resolution, prolonged IgM elevation, and increased complement activity. Conclusions: One year after hospitalization, most patients achieved substantial clinical improvement, but immune reconstitution lagged behind. These findings highlight the dissociation between clinical and immunological recovery and suggest that persistent immune dysregulation may be associated with long COVID manifestations. Incorporating immune monitoring into post-COVID care may help identify patients at risk of prolonged sequelae and guide targeted therapeutic strategies.
Full article
(This article belongs to the Special Issue Coronavirus Pathogenesis and Virus-Host Interaction)
►▼
Show Figures
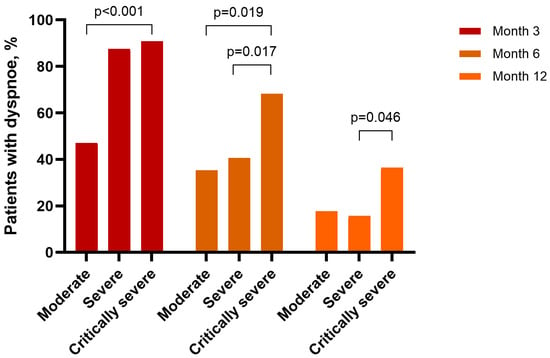
Figure 1
Open AccessOpinion
The Complexity of Bovine Leukemia Virus Oncogenesis
by
Florine Doucet, Alexis Fontaine, Malik Hamaidia, Jean-Rock Jacques, Thomas Jouant, Nour Mhaidly, Songkang Qin, Roxane Terres, Xavier Saintmard, Luc Willems and Manon Zwaenepoel
Viruses 2025, 17(12), 1609; https://doi.org/10.3390/v17121609 - 12 Dec 2025
Abstract
Bovine leukemia virus (BLV) is a retrovirus infecting several bovid species, notably Bos taurus, where it fulfills Koch’s postulates for pathogenicity. The virus primarily targets B-lymphocytes, establishing lifelong infections that remain mostly asymptomatic but can progress to lymphocytosis or lymphoma. Transmission occurs
[...] Read more.
Bovine leukemia virus (BLV) is a retrovirus infecting several bovid species, notably Bos taurus, where it fulfills Koch’s postulates for pathogenicity. The virus primarily targets B-lymphocytes, establishing lifelong infections that remain mostly asymptomatic but can progress to lymphocytosis or lymphoma. Transmission occurs through live infected cells via blood, milk, or transplacental routes. Despite a robust antiviral immunity, BLV replicates by producing virions (i.e., the infectious cycle) or inducing mitosis of infected cells (i.e., clonal expansion). The immune system effectively controls the infectious cycle but fails to impede clonal expansion, leading to chronic immune activation and immunosuppression. BLV modifies the transcriptome of the host cell by expressing oncogenic factors (Tax), viral microRNAs and antisense RNAs. Leukemogenesis arises from cumulative alterations of the virus (e.g., 5′-end deletions of the integrated provirus and histone modifications of the LTR promoter) and the host cell (e.g., genomic mutations and favorable chromatin integration). This model underscores a unique persistence strategy, linking chronic infection, immune evasion, and slow multistep oncogenesis in the bovine host.
Full article
(This article belongs to the Special Issue Human T-Cell Leukemia Virus (HTLV) Infection and Treatment: 2nd Edition)
►▼
Show Figures

Graphical abstract
Open AccessArticle
Long-Term Transcriptomic Reprogramming in Peripheral Blood Mononuclear Cells of Severe COVID-19 Survivors Reveals Pro-Oncogenic Signatures and Cancer-Associated Hub Genes
by
Pelin Duru Cetinkaya, Ozgecan Kayalar, Vahap Eldem, Serap Argun Baris, Nurdan Kokturk, Selim Can Kuralay, Hadi Rajabi, Nur Konyalilar, Deniz Mortazavi, Seval Kubra Korkunc, Sinem Erkan, Gizem Tuse Aksoy, Gul Eyikudamaci, Pelin Pinar Deniz, Oya Baydar Toprak, Pinar Yildiz Gulhan, Gulseren Sagcan, Neslihan Kose Kabil, Aysegul Tomruk Erdem, Fusun Fakili, Onder Ozturk, Ilknur Basyigit, Hasim Boyaci, Emel Azak, Tansu Ulukavak Ciftci, Ipek Kivilcim Oguzulgen, Hasan Selcuk Ozger, Pinar Aysert Yildiz, Ismail Hanta, Ozlem Ataoglu, Merve Ercelik, Caglar Cuhadaroglu, Hacer Kuzu Okur, Muge Meltem Tor, Esra Nurlu Temel, Seval Kul, Yıldız Tutuncu, Oya Itil and Hasan Bayramadd
Show full author list
remove
Hide full author list
Viruses 2025, 17(12), 1608; https://doi.org/10.3390/v17121608 - 12 Dec 2025
Abstract
This study examined the long-term transcriptomic reprogramming in peripheral blood mononuclear cells (PBMCs) of severe COVID-19 patients and its effects for cancer development. RNA sequencing was conducted on PBMCs obtained from healthy controls, COVID-19 patients without pneumonia, and COVID-19 patients exhibiting severe pneumonia
[...] Read more.
This study examined the long-term transcriptomic reprogramming in peripheral blood mononuclear cells (PBMCs) of severe COVID-19 patients and its effects for cancer development. RNA sequencing was conducted on PBMCs obtained from healthy controls, COVID-19 patients without pneumonia, and COVID-19 patients exhibiting severe pneumonia one year post-infection. Differential gene expression analysis identified a sustained pro-oncogenic molecular signature, especially among severe COVID-19 patients. Functional enrichment analysis revealed a substantial enrichment of cancer-associated pathways, encompassing apoptosis, viral carcinogenesis, and transcriptional dysregulation. Notably, the autophagy-related gene SQSTM1/P62 was recognized as a distinctive hub gene within the severe COVID-19 patients, interacting with pivotal genes associated with inflammation, apoptosis, and cancer advancement. Survival analysis demonstrated that elevated expression of COVID-19-associated hub genes correlated with unfavorable prognosis in various cancer types, including adrenocortical carcinoma, bladder urothelial carcinoma, and brain lower-grade glioma. These findings indicate that severe COVID-19 infection may establish a systemic milieu favorable to cancer development or recurrence, highlighting the necessity of prolonged oncological monitoring in these patients. Finding specific molecular targets and pathways can help us understand how COVID-19 might be linked to a higher risk of cancer.
Full article
(This article belongs to the Special Issue Beyond Acute: Navigating Long COVID and Post-Viral Complications)
►▼
Show Figures

Figure 1
Open AccessArticle
Efficacy of a Modified-Live Virus Combination Vaccine (CDV, CAV, CPV, CPiV), CanigenTM DHPPi, in Puppies Vaccinated at Six Weeks of Age
by
Sofia Loukeri, Fabien Senseby, Elodie Benizeau, Joelle Cronier and Sylvie Gueguen
Viruses 2025, 17(12), 1607; https://doi.org/10.3390/v17121607 - 12 Dec 2025
Abstract
During their early life, puppies are protected against infectious agents with the presence of maternal derived antibodies (MDA). Vaccination is recommended to start as soon as the levels of MDA begin to wane to ensure that the puppy’s immune system can respond effectively
[...] Read more.
During their early life, puppies are protected against infectious agents with the presence of maternal derived antibodies (MDA). Vaccination is recommended to start as soon as the levels of MDA begin to wane to ensure that the puppy’s immune system can respond effectively to the vaccines and develop active immunity against diseases. Two studies were designed to assess the efficacy of the CanigenTM DHPPi vaccine in puppies from 6 weeks of age. The studies comprised two parts: the efficacy assessment of the Canine Parainfluenza Virus (CPiV) vaccine component against a virulent challenge with CPiV (Experiment A) and an immunogenicity assessment of Canine Distemper Virus (CDV), Canine Adenovirus (CAV-2), and Canine Parvovirus (CPV-2) vaccine components (Experiment B). In Experiment A, twelve puppies were immunized with two injections of CanigenTM DHPPi (at minimum titer) two weeks apart and twelve control puppies received the vaccine diluent. All animals were challenged with a virulent, heterologous CPiV strain two weeks after the second vaccination. Vaccinated puppies exhibited a significant reduction in nasal viral shedding compared to the control group. Clinical signs of respiratory disease were mild and transient in both groups. In Experiment B, six puppies were immunized with two injections of CanigenTM DHPPi (at minimum titer) two weeks apart. A follow-up of the seroneutralizing antibodies titers against the CDV, CAV-2 and CPV-2 vaccine components was performed in order to assess the efficacy on the serological response basis. After the first vaccine injection, all the puppies seroconverted and presented seroneutralizing antibody titers reaching a protective thresholds against CDV (≥100.82), CAV-2 and CAV-1 (≥100.82), CPV-2 and CPV-2c (≥101.8). After the second vaccine injection, a more robust immune response was observed and the seroneutralizing antibody titers remained high until 4 weeks post vaccination for those three vaccine components. In both experiments (A and B), all vaccinated animals remained in good health, with no adverse reactions recorded during the vaccination phase. As a conclusion, the efficacy of the CanigenTM DHPPi vaccine was demonstrated when administered in dogs from 6 weeks of age. These results fully support the interest of an early vaccination in such young puppies followed by the recommended vaccination scheme.
Full article
(This article belongs to the Section Viral Immunology, Vaccines, and Antivirals)
►▼
Show Figures
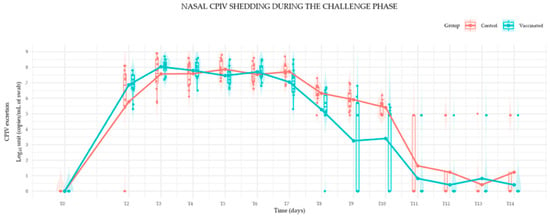
Figure 1
Open AccessArticle
Identification and Characterization of a Novel BVDV-1b Cluster in Sardinia Through Whole Genome Sequencing
by
Giada Lostia, Elisabetta Coradduzza, Loris Bertoldi, Angela Maria Rocchigiani, Roberto Bechere, Cinzia Pasini, Lorenzo Stevanato, Mariangela Stefania Fiori, Angelo Ruiu and Giantonella Puggioni
Viruses 2025, 17(12), 1606; https://doi.org/10.3390/v17121606 - 12 Dec 2025
Abstract
Bovine viral diarrhea (BVD) is a highly infectious disease with a global distribution caused by the bovine viral diarrhea virus (BVDV), primarily affecting cattle. Dairy farms play a central role in the persistence and spread of BVDV in Italy, making control strategies and
[...] Read more.
Bovine viral diarrhea (BVD) is a highly infectious disease with a global distribution caused by the bovine viral diarrhea virus (BVDV), primarily affecting cattle. Dairy farms play a central role in the persistence and spread of BVDV in Italy, making control strategies and genetic studies essential to reduce its circulation. This work aimed to identify and characterize the genotype and subgenotype of BVDV infecting cattle in a specific area of Sardinia. Ten BVDV Sardinian strains were isolated and sequenced from the blood of infected cattle collected into EDTA tubes during outbreaks between 2018 and 2024. Then, to characterize the isolates, phylogenetic and variant analyses were performed on the entire collection of BVDV genomes available to date in GenBank. All Sardinian isolates were assigned to the BVDV-1b subgenotype. Except for two divergent strains, the isolates clustered into a distinct monophyletic clade characterized by 61 exclusive variants absent in all other analyzed sequences. These findings point to the existence of a distinct Sardinian genomic signature. Moreover, among these mutations, 19 missenses distributed on genes encoding the E1, E2, Core, NS3, NS4B and NS5A proteins could have a relevant functional impact, given the role these proteins play in the virus life cycle and in interaction with the host.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
Figure 1
Open AccessRetraction
RETRACTED: Kulanayake et al. Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins. Viruses 2024, 16, 732
by
Shermila Kulanayake, Faryal Dar and Suresh K. Tikoo
Viruses 2025, 17(12), 1605; https://doi.org/10.3390/v17121605 - 12 Dec 2025
Abstract
The journal retracts the article, “Regions of Bovine Adenovirus-3 Protein VII Involved in Interactions with Viral and Cellular Proteins” [...]
Full article
(This article belongs to the Section Animal Viruses)
Open AccessArticle
High-Fidelity and Cost-Effective Engineering of SARS-CoV-2
by
Marco Olguin-Nava, Thomas Hennig, Charlene Börtlein, Patrick Bohn, Uddhav B. Ambi, Alexander Gabel, Lina M. Günter, Anne-Sophie Gribling-Burrer, Nora Schmidt, Neva Caliskan, Lars Dölken, Mathias Munschauer and Redmond P. Smyth
Viruses 2025, 17(12), 1604; https://doi.org/10.3390/v17121604 - 11 Dec 2025
Abstract
Efficient reverse genetics systems are essential for understanding SARS-CoV-2 pathogenesis, host–virus interactions, and potential therapeutic interventions. Here, we developed a cost-effective PCR-based reverse genetics platform that splits the SARS-CoV-2 genome into only six bacterial plasmids, enabling cloning, manipulation, and the rescue of recombinant
[...] Read more.
Efficient reverse genetics systems are essential for understanding SARS-CoV-2 pathogenesis, host–virus interactions, and potential therapeutic interventions. Here, we developed a cost-effective PCR-based reverse genetics platform that splits the SARS-CoV-2 genome into only six bacterial plasmids, enabling cloning, manipulation, and the rescue of recombinant SARS-CoV-2 (rSARS-CoV-2) with high fidelity and high viral titers after a single passage. Using this system, we generated and characterized spike protein mutants Y453F and N501Y, as well as a U76G mutation in the 5′-UTR. Y453F showed reduced replication kinetics, lower cell binding, and diminished fitness, while N501Y exhibited comparable replication and fitness, highlighting the distinct effects of these spike protein mutations. The U76G mutation is located within a novel NSP9 binding site in the 5′-UTR and leads to impaired RNA synthesis and reduced viral replication efficiency, suggesting an important role in transcription and replication. Our findings highlight the robustness and adaptability of this reverse genetics system, providing a versatile, cost-effective tool for studying SARS-CoV-2 mutations and their effects on replication and fitness, with potential applications in vaccine and therapeutic development.
Full article
(This article belongs to the Special Issue Viral RNA and Its Interaction with the Host)
►▼
Show Figures
Figure 1
Open AccessConference Report
First Brazilian Symposium on Viruses of Microorganisms (BrVoM 2025)
by
Jônatas Santos Abrahão, Luiz Felipe Leomil Coelho, Amanda Stéphanie Arantes Witt, Ana Karoline da Nóbrega Nunes Alves, Anna Catarina Dias Soares Guimarães, Bárbara Stehling Ramos Silva, Bruna Nascimento Neiva, Bruno Fernandes de Oliveira, Jamile Dias, João Victor Rodrigues Pessoa Carvalho, Letícia Pereira Lopes, Matheus Felipe dos Reis Rodrigues, Matheus Gomes Barcelos, Nidia Esther Colquehuanca Arias, Poliane Zerbini, Vera Lucia dos Santos, Caio Ambrosio Leal-Dutra, Savio Torres de Farias, Rodrigo Araujo Lima Rodrigues, Juliana Reis Cortines, Otavio Henrique Thiemann, Paulo Boratto, Marcelo Henrique Aguiar de Freitas and Gabriel Magno de Freitas Almeidaadd
Show full author list
remove
Hide full author list
Viruses 2025, 17(12), 1603; https://doi.org/10.3390/v17121603 - 11 Dec 2025
Abstract
In recent decades, there has been an increased interest in viruses of microorganisms (VoM) and international efforts to gather researchers interested in them. Here, we describe the 1st Brazilian Symposium on Viruses of Microorganisms (BrVoM), held on 1 August 2025 at the Federal
[...] Read more.
In recent decades, there has been an increased interest in viruses of microorganisms (VoM) and international efforts to gather researchers interested in them. Here, we describe the 1st Brazilian Symposium on Viruses of Microorganisms (BrVoM), held on 1 August 2025 at the Federal University of Minas Gerais (UFMG, Belo Horizonte, Brazil) with institutional support from the Federal University of Alfenas (UNIFAL) and the Brazilian Society for Virology (SBV). The symposium greatly surpassed expectations, gathering nearly 300 attendees from all Brazilian geographical regions. The scientific program included keynote and thematic lectures covering bacteriophages, fungal viruses, giant viruses, and microbial resources regulation. The event was remarkable for its collaborative spirit and inclusion of early career attendees. The success of this first edition highlights the vitality of the Brazilian community working on microbial viruses and sets the stage for future editions.
Full article
(This article belongs to the Section General Virology)
►▼
Show Figures

Figure 1
Open AccessArticle
Hepatitis C Virus Infection in Mongolia: Updated Provincial Data on Prevalence, Genotype Distribution, and Age-Specific Risk Factors
by
Amgalan Byambasuren, Myagmarjaltsan Baatarzorigt, Munkhtuya Otgon, Byambasuren Bat-Amgalan, Mandakhnaran Purevkhuu, Naranzul Nyamsuren, Enkh-Amar Ayush, Dashchirev Munkh-Orshikh, Khurelbaatar Nyamdavaa and Oidov Baatarkhuu
Viruses 2025, 17(12), 1602; https://doi.org/10.3390/v17121602 - 11 Dec 2025
Abstract
(1) Background: Mongolia has historically reported one of the highest hepatitis C virus (HCV) prevalence rates globally, with past national estimates exceeding 15%, making HCV infection a major public health priority. This study aimed to assess the prevalence, genotype distribution, and risk factors
[...] Read more.
(1) Background: Mongolia has historically reported one of the highest hepatitis C virus (HCV) prevalence rates globally, with past national estimates exceeding 15%, making HCV infection a major public health priority. This study aimed to assess the prevalence, genotype distribution, and risk factors of HCV infection among residents of Arkhangai Province. (2) Methods: This population-based cross-sectional study was conducted in 2022 including 2304 individuals aged 0–80 years. Serum samples were tested for anti-HCV antibodies using ELISA and for HCV RNA using PCR. Positive samples were genotyped, and demographic and exposure data were analyzed using logistic regression to identify independent risk factors. (3) Results: The prevalence of anti-HCV antibodies was 12.0%, and HCV RNA positivity was 7.16%. Infection increased significantly with age (p < 0.001) and was higher among females (14.6%) than males (8.4%). Genotype 1b predominated (98.2%), followed by 1a (1.2%) and 2 (0.6%). Several exposures showed strong associations with HCV infection in univariate analysis, including cupping therapy (OR 2.37, 95% CI 1.71–3.28), shared razor use (OR 2.39, 95% CI 1.59–3.60), cosmetic procedures (OR 1.70, 95% CI 1.11–2.45), and unsafe injections (OR 2.06, 95% CI 1.40–3.02). In multivariable analysis, four exposures remained independently associated with HCV infection: cupping therapy (adjusted OR 1.89, 95% CI 1.32–2.70), shared razor use (adjusted OR 1.98, 95% CI 1.24–2.89), cosmetic procedures (adjusted OR 1.62, 95% CI 1.39–2.24), and unsafe injections (adjusted OR 1.84, 95% CI 1.19–2.83). (4) Conclusions: HCV infection remains prevalent, particularly among older adults and women. Genotype 1b continues to predominate, indicating that the viral genetic distribution has remained largely unchanged over the past decade. Continued education, safe injection practices, and regulation of traditional and cosmetic procedures are essential to reduce HCV transmission and support Mongolia’s elimination goals. These findings highlight the need for comprehensive prevention strategies addressing both unsafe traditional/medical practices and the rapidly expanding cosmetic and aesthetic service sector.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
►▼
Show Figures

Figure 1
Open AccessEditorial
Viral Sepsis: Pathogenesis, Diagnostics and Therapeutics—A Summary of Key Findings and Insights from the Special Issue
by
Stamatia Tsoupra and Karolina Akinosoglou
Viruses 2025, 17(12), 1601; https://doi.org/10.3390/v17121601 - 10 Dec 2025
Abstract
Viral sepsis has gained increasing clinical and scientific attention as it represents a complex, life-threatening condition driven by a dysregulated host response to viral pathogens [...]
Full article
(This article belongs to the Special Issue Viral Sepsis: Pathogenesis, Diagnostics and Therapeutics)
Open AccessArticle
Characterization of Novel Przondovirus Phage Adeo Infecting Klebsiella pneumoniae of the K39 Capsular Type
by
Nadezhda V. Kolupaeva, Peter V. Evseev, Victoria A. Avdeeva, Angelika A. Sizova, Natalia E. Suzina, Nikolay V. Volozhantsev and Anastasia V. Popova
Viruses 2025, 17(12), 1600; https://doi.org/10.3390/v17121600 - 10 Dec 2025
Abstract
Klebsiella pneumoniae is one of the most significant nosocomial pathogens and an important cause of human infections worldwide. The microorganism is capable of producing different capsular polysaccharides (CPSs), which are the primary receptors for capsule-specific K. pneumoniae bacteriophages encoding tailspike proteins (TSPs) with
[...] Read more.
Klebsiella pneumoniae is one of the most significant nosocomial pathogens and an important cause of human infections worldwide. The microorganism is capable of producing different capsular polysaccharides (CPSs), which are the primary receptors for capsule-specific K. pneumoniae bacteriophages encoding tailspike proteins (TSPs) with polysaccharide-degrading activity. In this study, the novel virulent Przondovirus phage Adeo was isolated and characterized. The phage was able to infect K. pneumoniae strain with a K39 capsular polysaccharide structure. The morphology, biological properties, stability, and genomic organization of Adeo were studied. Comparative genomic and phylogenetic analyses were performed to establish the relationship between the phage and other bacterial viruses. The gene encoding TSP Adeo_gp48 was identified and cloned. Recombinant depolymerase lacking the N-terminal part was expressed, purified, and formed an opaque zone of CPS depolymerization on the K39 K. pneumoniae bacterial lawns. The structural and phylogenetic similarities of Adeo’s TSP to other phage-encoded depolymerases were discussed.
Full article
(This article belongs to the Section Bacterial Viruses)
►▼
Show Figures
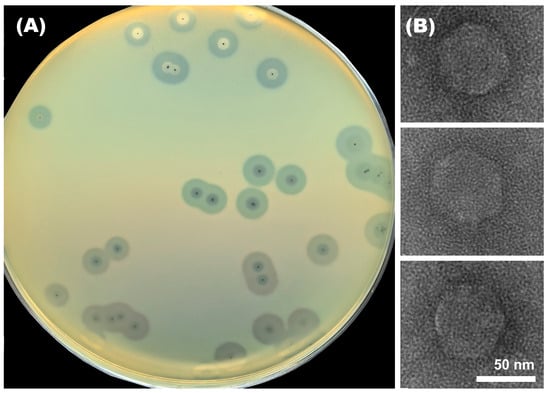
Figure 1
Open AccessArticle
The Intrinsic Innate Immunity of Hepatocytes Suppresses HBV Replication and Is Antagonized by HBx
by
Chui Zeng, Fayed Attia Koutb Megahed, Yiqiong Guo, Dongmei Sun, Yaru Wang, Qin Liu, Yanwei Bi, Jinghang Li, Qi Zhou, Qingdong Xie, Pingnan Sun and Xiaoling Zhou
Viruses 2025, 17(12), 1599; https://doi.org/10.3390/v17121599 - 10 Dec 2025
Abstract
(1) Background: Hepatitis B virus (HBV) belongs to the Hepadnaviridae family of viruses that interact with hepatocytes. HBV infection is a major global health problem. Most adults clear the infection quickly after being infected with HBV, while a few people develop chronic HBV
[...] Read more.
(1) Background: Hepatitis B virus (HBV) belongs to the Hepadnaviridae family of viruses that interact with hepatocytes. HBV infection is a major global health problem. Most adults clear the infection quickly after being infected with HBV, while a few people develop chronic HBV infection. It is well-known that the early innate immune response of host cells plays an important role in the fight against virus infection. However, the interactions between HBV and the intrinsic innate immune system of hepatocytes are still not fully understood. The aim of this study was to confirm the interaction between HBV and hepatocytes, and to identify the interferon-stimulated genes (ISGs) regulated by HBx and their expression in association with HBV-associated HCC (HBV-HCC), so that we can refine our understanding of the interaction between HBV and ISGs and its potential influence on HBV-HCC. (2) Methods: We analyzed data concerning the stimulation of IFN-dependent genes in primary human hepatocytes (PHHs) transfected with pathogen DNA mimetics or infected with HBV in the GSE69590 database. Bioinformatic methods, such as GSEA, GO, and KEGG, were used to analyze the differentially expressed innate immunity genes and their related pathways to identify candidate intrinsic innate immune factors. qPCR on HepG2 and Huh7 cells, which highly express HBx, was used to detect relevant intrinsic innate immune factors. qPCR, RNAi, and Elisa methods were used to identify intrinsic innate immune factors in HBV-integrated HepG2.2.15 cells, and bioinformatics analysis was conducted on the HBV-infected tissues and cells in the GEO database. (3) Results: Inhibition of the JAK-STAT pathway enhanced HBV replication in HepG2 cells transfected with HBV plasmid and HepG2-NTCP cells infected with HBV. GSEA analysis of the GSE69590 data revealed significant changes in intrinsic innate immune pathways during HBV infection with PHH for 40 h. A total of 84 differentially expressed, candidate innate immunity genes were identified in GSE69590. Validation showed that TRIM22 and TRIM56 were down-regulated when HBx was expressed. Consistently, TRIM22 and TRIM56 were up-regulated following inhibition of HBx by transfection of HBx siRNA into HepG2.2.15 cells, and HBV pgRNA was up-regulated following down-regulated expression of TRIM22 and TRIM56 in HEK293 cells. Receiver operating characteristics (ROC) and overall survival (OS) analysis of 204 HBV-HCC patients showed that expression of TRIM22 was closely associated with HBV-HCC, and high expression of TRIM22 was associated with longer survival. (4) Conclusions: Innate immunity genes TRIM22 and TRIM56 are regulated by HBx, and higher expression of TRIM22 is closely related to longer survival of HBV-HCC patients.
Full article
(This article belongs to the Special Issue Interferon-Stimulated Genes in Antiviral Immunity)
►▼
Show Figures
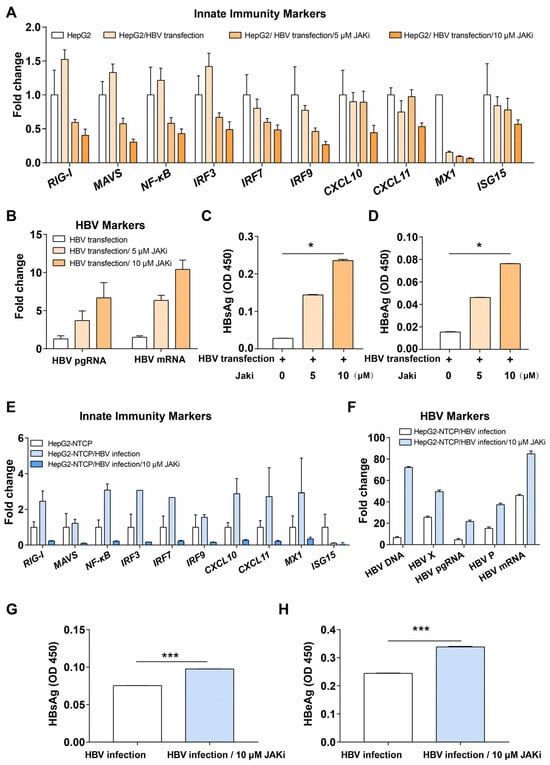
Figure 1
Open AccessArticle
Nowcast-It: A Practical Toolbox for Real-Time Adjustment of Reporting Delays in Epidemic Surveillance
by
Amna Tariq, Ping Yan, Amanda Bleichrodt and Gerardo Chowell
Viruses 2025, 17(12), 1598; https://doi.org/10.3390/v17121598 - 10 Dec 2025
Abstract
One difficulty that arises in tracking and forecasting real-time epidemics is reporting delays, which are defined as the inherent delay between the time of symptom onset and the time a case is reported. Reporting delays can be caused by delays in case detection,
[...] Read more.
One difficulty that arises in tracking and forecasting real-time epidemics is reporting delays, which are defined as the inherent delay between the time of symptom onset and the time a case is reported. Reporting delays can be caused by delays in case detection, symptom onset after infection, seeking medical care, or diagnostics, and they distort the accurate forecasting of diseases during epidemics and pandemics. To address this, we introduce a practical nowcasting approach grounded in survival analysis and actuarial science, explicitly allowing for non-stationarity in reporting delay patterns to better capture real-world variability. Despite its relevance, no flexible and accessible toolbox currently exists for non-stationary delay adjustment. Here, we present Nowcast-It, a tutorial-based toolbox that includes two components: (1) an R code base for delay adjustment and (2) a user-friendly R-Shiny application to enable interactive visualization and reporting delay correction without prior coding expertise. The toolbox supports daily, weekly, or monthly resolution data and enables model performance assessment using metrics such as mean absolute error, mean squared error, and 95% prediction interval coverage. We demonstrate the utility of Nowcast-It toolbox using publicly available weekly Ebola case data from the Democratic Republic of Congo. We and others have adjusted for reporting delays in real-time analyses (e.g., Singapore) and produced early COVID-19 forecasts; here, we package those delay adjustment routines into an accessible toolbox. It is designed for researchers, students, and policymakers alike, offering a scalable and accessible solution for addressing reporting delays during outbreaks.
Full article
(This article belongs to the Special Issue Multiscale Modeling and Forecasting of COVID-19 and Respiratory Virus Dynamics)
►▼
Show Figures

Figure 1
Open AccessArticle
Raspberry Viruses in the Czech Republic, with Identification of a Novel Virus: Raspberry Virus A
by
Jiunn Luh Tan, Igor Koloniuk, Ondřej Lenz, Jana Veselá, Jaroslava Přibylová, Rostislav Zemek, Josef Špak, Radek Čmejla, Jiří Sedlák, Dag-Ragnar Blystad, Zhibo Hamborg and Jana Fránová
Viruses 2025, 17(12), 1597; https://doi.org/10.3390/v17121597 - 9 Dec 2025
Abstract
Although global raspberries production has grown in the past decade, it remains threatened by plant viruses. This study surveyed raspberry viruses and associated arthropods in the Czech Republic between 2021 and 2022 across five regions. A total of 257 plant and 151 arthropod
[...] Read more.
Although global raspberries production has grown in the past decade, it remains threatened by plant viruses. This study surveyed raspberry viruses and associated arthropods in the Czech Republic between 2021 and 2022 across five regions. A total of 257 plant and 151 arthropod samples were tested using RT-(q)PCR for 12 viruses listed in the EPPO Certification scheme, plus raspberry leaf blotch virus (RLBV) and a novel virus, tentatively named raspberry-associated virus A (RaVA). Raspberry bushy dwarf virus (RBDV) was most prevalent (51.8%), followed by black raspberry necrosis virus (BRNV, 42.0%) and raspberry leaf mottle virus (RLMV, 28.4%). Four viruses—arabis mosaic virus, apple mosaic virus, strawberry latent ringspot virus, raspberry ringspot virus—were not detected. RBDV was also identified in Sambucus nigra, a new host, while mixed RLBV and RaVA infection was found in wild Rubus occidentalis. RLBV was experimentally transmitted to Nicotiana occidentalis 37B in the presence of Phyllocoptes gracilis. Seven of 39 arthropod species carried viruses, but only two—Amphorophora rubi idaei and Aphis idaei—are known vectors. PCR amplicons from 92 isolates were sequenced, revealing high variability in several viruses. These findings offer new insights but highlight the need for continued monitoring and research.
Full article
(This article belongs to the Section Viruses of Plants, Fungi and Protozoa)
►▼
Show Figures
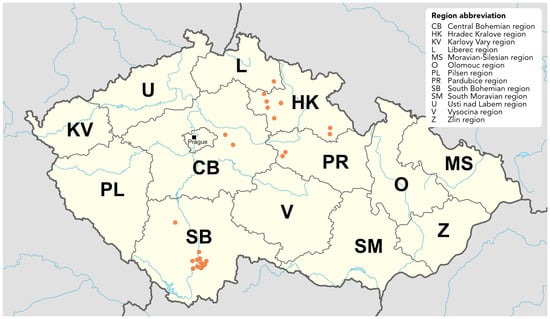
Figure 1

Journal Menu
► ▼ Journal Menu-
- Viruses Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
Viruses
Structure-Based Antiviral Drugs and Vaccine Design
Guest Editors: Shi-hua Xiang, Fengwei BaiDeadline: 15 December 2025
Special Issue in
Viruses
Bioinformatics and Computational Approaches in Viral Genomics and Evolution 2025
Guest Editors: Camila Romano, Mariana Severo Ramundo, Nilson CoimbraDeadline: 16 December 2025
Special Issue in
Viruses
Virology in Italy 2025—9th National Congress of the Italian Society for Virology
Guest Editors: Enzo Tramontano, Arnaldo Caruso, Massimiliano Galdiero, Luisa Rubino, Gabriele VaccariDeadline: 20 December 2025
Special Issue in
Viruses
Diversity and Coinfections of Plant or Fungal Viruses, 3rd Edition
Guest Editors: Islam Hamim, Ken Komatsu, Hiromitsu MoriyamaDeadline: 20 December 2025
Topical Collections
Topical Collection in
Viruses
Poxviruses
Collection Editors: Giliane de Souza Trindade, Galileu Barbosa Costa, Flavio Guimaraes da Fonseca
Topical Collection in
Viruses
Efficacy and Safety of Antiviral Therapy
Collection Editors: Giordano Madeddu, Andrea De Vito, Agnese Colpani
Topical Collection in
Viruses
Phage Therapy
Collection Editors: Nina Chanishvili, Jean-Paul Pirnay, Mikael Skurnik









