- Article
Morphological and Immunohistochemical Characteristics of Liver Inflammation in Patients with a History of COVID-19
- Ilze Strumfa,
- Ludmila Viksna and
- Oksana Kolesova
- + 6 authors
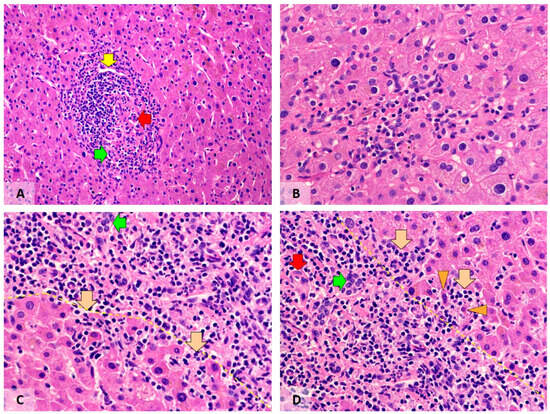
The COVID-19 pandemic caused more than seven million deaths, mostly via acute respiratory distress syndrome with microvascular thrombosis. Compared to the amount of information about pulmonary pathology, information about COVID-19-induced liver lesions is scarce, especially with regard to the long-term consequences. The aim of our study was to evaluate inflammatory, vascular and fibrotic changes in hepatobiliary tissues of patients with a history of COVID-19 (post-COVID-19 patients). Based on the Knodell score, moderate portal inflammation was observed in 41.2% of post-COVID-19 patients, contrasting with 14.3% of control cases (p = 0.06). Moderate periportal inflammation was present in 26.5% and 7.1% of patients, respectively (p = 0.08). Post-COVID-19 patients showed higher counts of CD3+ lymphocytes (p = 0.02) and lower counts of CD68+ macrophages (p = 0.04), as well as more frequent and extensive regenerative changes in hepatocytes and the biliary epithelium (p = 0.0007). We did not find significant fibrosis or pathological changes in blood vessels, and only mild steatosis was observed in both groups.
2 January 2026