The ORF1ab of Feline Coronavirus Plays a Critical Role in Regulating the Innate Immune Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Antibodies
2.2. Rescue of the Recombinant Virus
2.3. Isolation and Culture of Feline BMDMs
2.4. Viral Growth Kinetics
2.5. Immunofluorescence Assay
2.6. Electron Microscopy Identification
2.7. Analysis of Gene Expression Using RT-qPCR
2.8. Dual-Luciferase Reporter Assay
2.9. Transcriptomic Assay
2.10. Proteomic Assay
2.11. Western Blotting
2.12. Plasmid Construction
3. Results
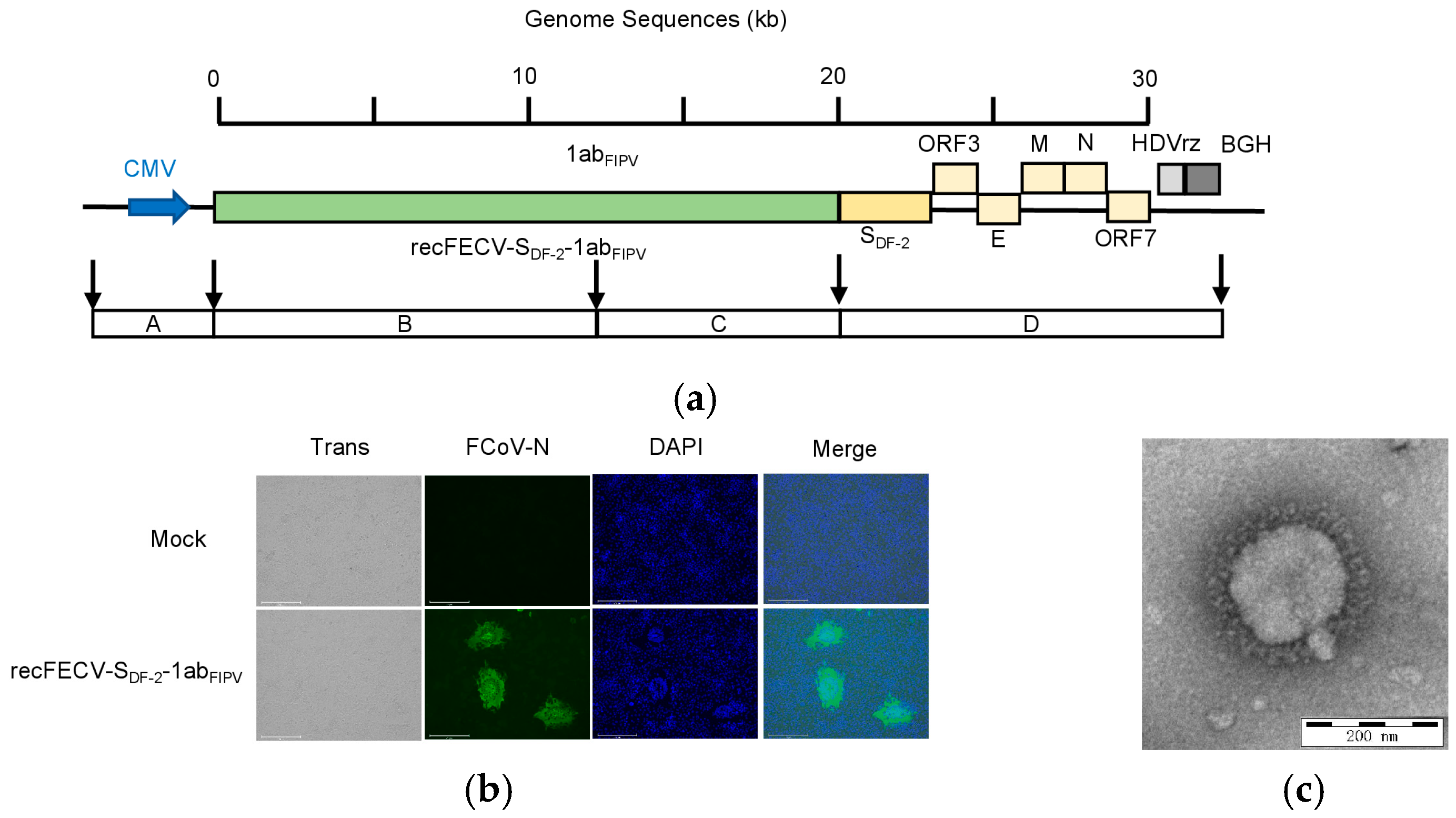
3.1. Rescue of the recFECV-SDF-2-1abFIPV
3.2. Growth Kinetics of the recFECV-SDF-2-1abFIPV
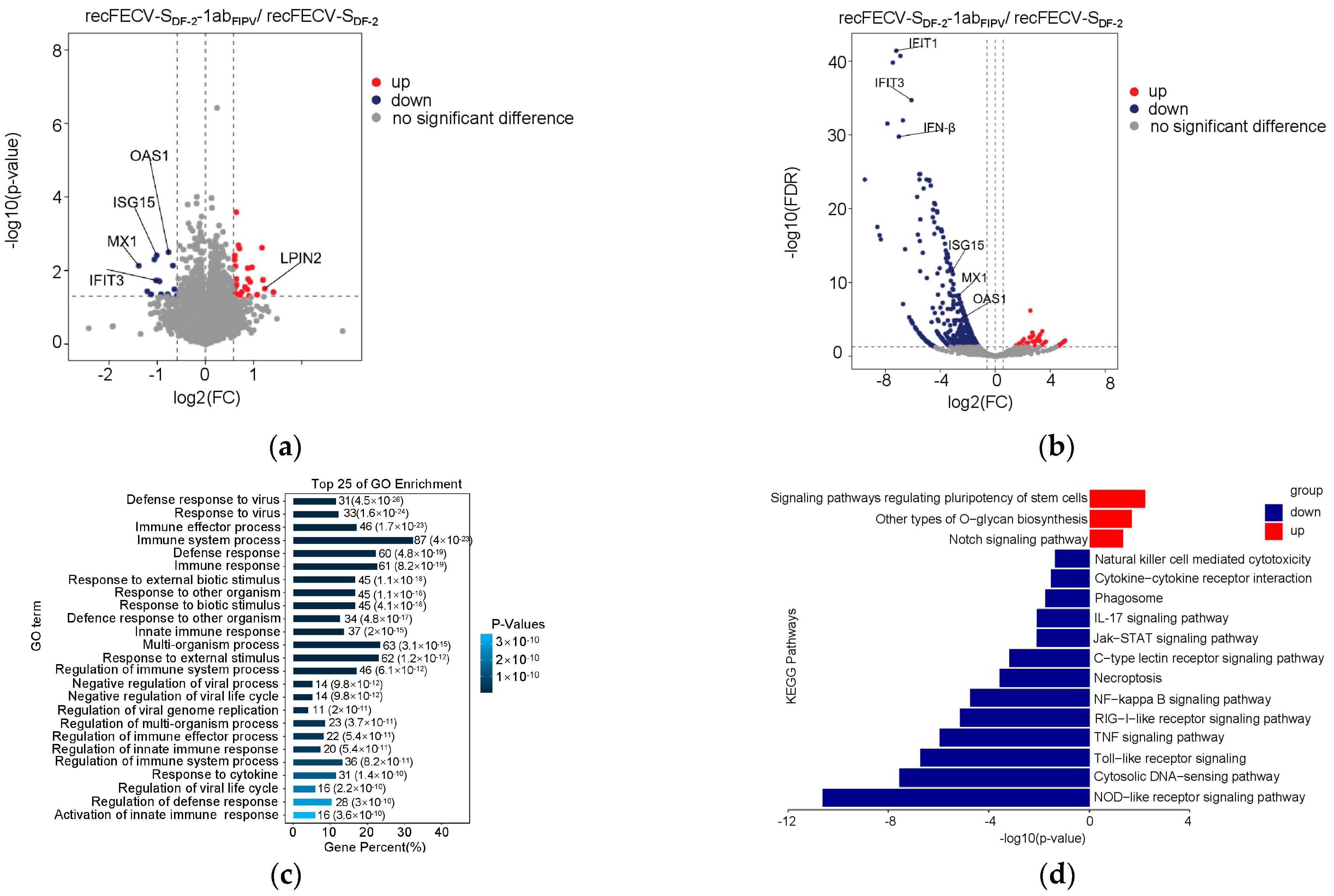
3.3. Infection of recFECV-SDF-2-1abFIPV Causes Profound Changes in Protein Expression in CRFK Cells
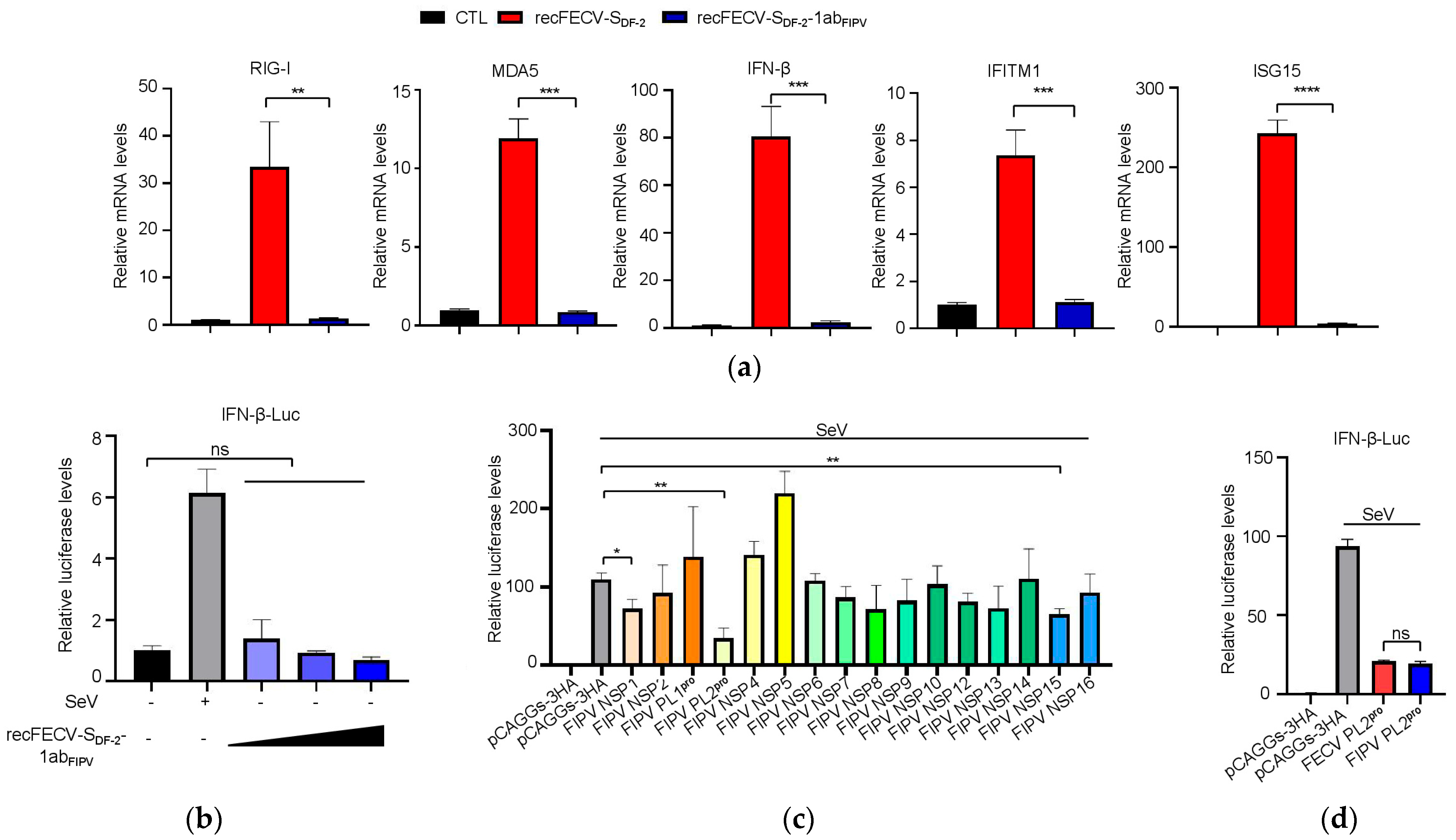
3.4. The Presence of ORF1ab of FIPV in the Recombinant Virus Enhances Innate Immune Suppression
3.5. NSP1 and NSP3 Are Involved in the Function of Innate Immune Suppression
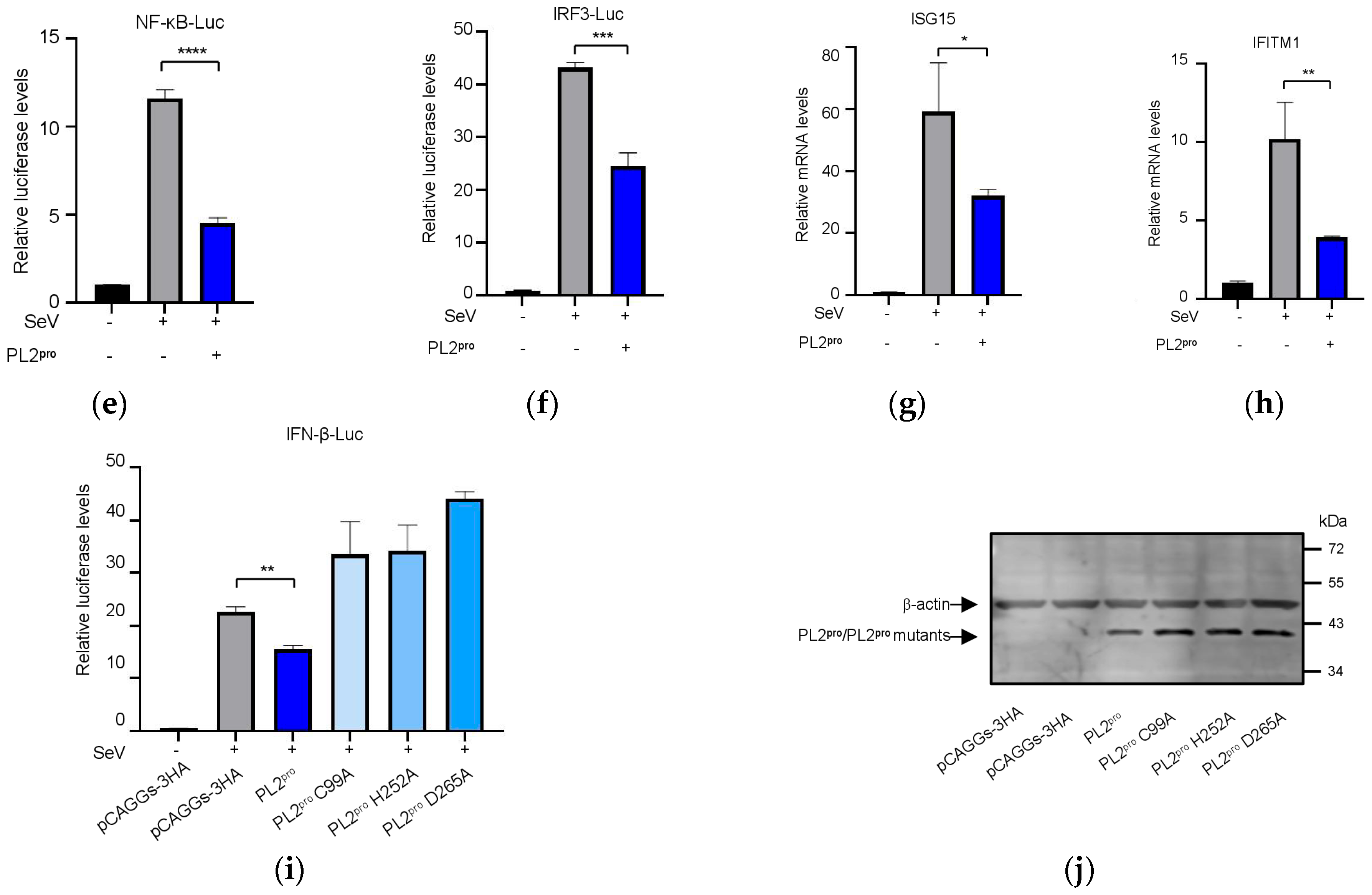
3.6. The PL2 Protease Activity of NSP3 Is Required for Its Role in Innate Immune Suppression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FIP | Feline infectious peritonitis |
| FIPV | Feline infectious peritonitis virus |
| FECV | Feline enteric coronavirus |
| FCoV | Feline coronavirus |
| IFN-β | Interferon β |
| S | Spike protein |
| E | Envelope protein |
| M | Membrane protein |
| N | Nucleocapsid protein |
| PL2 | Papain-like protease 2 |
| CRFK | Crandell feline kidney |
| Fcwf-4 | Felis catus whole fetus-4 |
| Luc | Luciferase |
| SeV | Sendai virus |
| NSPs | Non-structural proteins |
| DMEM | Dulbecco’s modified Eagle’s medium |
| PBS | Phosphate-buffered saline |
| CPE | Cytopathic effects |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ISGs | Interferon-stimulated genes |
| FDR | False discovery rate |
| EMEM | Eagle’s minimum essential medium |
| RT-qPCR | Quantitative reverse transcription PCR |
References
- Evermann, J.F.; Baumgartener, L.; Ott, R.L.; Davis, E.V.; McKeirnan, A.J. Characterization of a feline infectious peritonitis virus isolate. Vet. Pathol. 1981, 18, 256–265. [Google Scholar] [CrossRef]
- Poland, A.M.; Vennema, H.; Foley, J.E.; Pedersen, N.C. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J. Clin. Microbiol. 1996, 34, 3180–3184. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Boyle, J.F.; Floyd, K.; Fudge, A.; Barker, J. An enteric coronavirus infection of cats and its relationship to feline infectious peritonitis. Am. J. Vet. Res. 1981, 42, 368–377. [Google Scholar] [CrossRef]
- Vennema, H.; Poland, A.; Foley, J.; Pedersen, N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology 1998, 243, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.E.; Poland, A.; Carlson, J.; Pedersen, N.C. Patterns of feline coronavirus infection and fecal shedding from cats in multiple-cat environments. J. Am. Vet. Med. Assoc. 1997, 210, 1307–1312. [Google Scholar] [CrossRef]
- Solikhah, T.I.; Agustin, Q.A.D.; Damaratri, R.A.; Siwi, D.A.F.; Rafi’uttaqi, G.N.; Hartadi, V.A.; Solikhah, G.P. A review of feline infectious peritonitis virus infection. Vet. World 2024, 17, 2417–2432. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, N.C. Virologic and immunologic aspects of feline infectious peritonitis virus infection. Adv. Exp. Med. Biol. 1987, 218, 529–550. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.T.; Su, B.L.; Hsieh, L.E.; Chueh, L.L. An outbreak of feline infectious peritonitis in a Taiwanese shelter: Epidemiologic and molecular evidence for horizontal transmission of a novel type II feline coronavirus. Vet. Res. 2013, 44, 57. [Google Scholar] [CrossRef] [PubMed]
- Attipa, C.; Warr, A.S.; Epaminondas, D.; O’Shea, M.; Hanton, A.J.; Fletcher, S.; Malbon, A.; Lyraki, M.; Hammond, R.; Hardas, A.; et al. Feline infectious peritonitis epizootic caused by a recombinant coronavirus. Nature 2025, 645, 228–234. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Choi, A.; Gibson, C.A.; Lee, J.; Brown, J.T.; Stewart, C.; Joshi, A.; Harari, S.; Willoughby, I.; Treichel, C.; et al. Loss of FCoV-23 spike domain 0 enhances fusogenicity and entry kinetics. Nature 2025, 645, 235–243. [Google Scholar] [CrossRef]
- Olarte-Castillo, X.A.; Frazier, L.E.; Gomes Noll, J.C.; Choi, A.; Whittaker, G.R. Rethinking the drivers of coronavirus virulence and pathogenesis; toward an understanding of the dynamic world of mutations, indels, and recombination within the alphacoronaviruses. mBio 2025, e01921-25. [Google Scholar] [CrossRef]
- Pedersen, N.C. A review of feline infectious peritonitis virus infection: 1963–2008. J. Feline Med. Surg. 2009, 11, 225–258. [Google Scholar] [CrossRef]
- Bank-Wolf, B.R.; Stallkamp, I.; Wiese, S.; Moritz, A.; Tekes, G.; Thiel, H.J. Mutations of 3c and spike protein genes correlate with the occurrence of feline infectious peritonitis. Vet. Microbiol. 2014, 173, 177–188. [Google Scholar] [CrossRef]
- Kennedy, M.; Boedeker, N.; Gibbs, P.; Kania, S. Deletions in the 7a ORF of feline coronavirus associated with an epidemic of feline infectious peritonitis. Vet. Microbiol. 2001, 81, 227–234. [Google Scholar] [CrossRef]
- Thiel, V.; Thiel, H.J.; Tekes, G. Tackling feline infectious peritonitis via reverse genetics. Bioengineered 2014, 5, 396–400. [Google Scholar] [CrossRef]
- Ehmann, R.; Kristen-Burmann, C.; Bank-Wolf, B.; König, M.; Herden, C.; Hain, T.; Thiel, H.J.; Ziebuhr, J.; Tekes, G. Reverse Genetics for Type I Feline Coronavirus Field Isolate To Study the Molecular Pathogenesis of Feline Infectious Peritonitis. mBio 2018, 9, e01422-18. [Google Scholar] [CrossRef]
- Wang, G.; Hu, G.; Liang, R.; Shi, J.; Qiu, X.; Yang, Y.; Jiao, Z.; Chen, Y.; Shen, Z.; Li, M.; et al. Establishment of Full-Length cDNA Clones and an Efficient Oral Infection Model for Feline Coronavirus in Cats. J. Virol. 2021, 95, e0074521. [Google Scholar] [CrossRef]
- Takano, T.; Yamada, S.; Doki, T.; Hohdatsu, T. Pathogenesis of oral type I feline infectious peritonitis virus (FIPV) infection: Antibody-dependent enhancement infection of cats with type I FIPV via the oral route. J. Vet. Med. Sci. 2019, 81, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Nakaguchi, M.; Doki, T.; Hohdatsu, T. Antibody-dependent enhancement of serotype II feline enteric coronavirus infection in primary feline monocytes. Arch. Virol. 2017, 162, 3339–3345. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Troyer, J.L.; Pecon-Slattery, J.; Roelke, M.E.; O’Brien, S.J. Genetics and pathogenesis of feline infectious peritonitis virus. Emerg. Infect. Dis. 2009, 15, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Healey, E.A.; Andre, N.M.; Miller, A.D.; Whittaker, G.R.; Berliner, E.A. Outbreak of feline infectious peritonitis (FIP) in shelter-housed cats: Molecular analysis of the feline coronavirus S1/S2 cleavage site consistent with a ‘circulating virulent-avirulent theory’ of FIP pathogenesis. JFMS Open Rep. 2022, 8, 20551169221074226. [Google Scholar] [CrossRef]
- Dedeurwaerder, A.; Desmarets, L.M.; Olyslaegers, D.A.J.; Vermeulen, B.L.; Dewerchin, H.L.; Nauwynck, H.J. The role of accessory proteins in the replication of feline infectious peritonitis virus in peripheral blood monocytes. Vet. Microbiol. 2013, 162, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, B.L.; Devriendt, B.; Olyslaegers, D.A.; Dedeurwaerder, A.; Desmarets, L.M.; Favoreel, H.W.; Dewerchin, H.L.; Nauwynck, H.J. Suppression of NK cells and regulatory T lymphocytes in cats naturally infected with feline infectious peritonitis virus. Vet. Microbiol. 2013, 164, 46–59. [Google Scholar] [CrossRef]
- Chen, S.; Tian, J.; Li, Z.; Kang, H.; Zhang, J.; Huang, J.; Yin, H.; Hu, X.; Qu, L. Feline Infectious Peritonitis Virus Nsp5 Inhibits Type I Interferon Production by Cleaving NEMO at Multiple Sites. Viruses 2019, 12, 43. [Google Scholar] [CrossRef]
- Doki, T.; Yabe, M.; Takano, T.; Hohdatsu, T. Differential induction of type I interferon by type I and type II feline coronaviruses in vitro. Res. Vet. Sci. 2018, 120, 57–62. [Google Scholar] [CrossRef]
- Shen, Z.; Yang, Y.; Yang, S.; Zhang, G.; Xiao, S.; Fu, Z.F.; Peng, G. Structural and Biological Basis of Alphacoronavirus nsp1 Associated with Host Proliferation and Immune Evasion. Viruses 2020, 12, 812. [Google Scholar] [CrossRef]
- Cao, H.; Gu, H.; Kang, H.; Jia, H. Development of a rapid reverse genetics system for feline coronavirus based on TAR cloning in yeast. Front. Microbiol. 2023, 14, 1141101. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Q.; Kong, F.; Guo, D.; Zhai, J.; Su, M.; Sun, D. Circulation and genetic diversity of Feline coronavirus type I and II from clinically healthy and FIP-suspected cats in China. Transbound. Emerg. Dis. 2019, 66, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Kouprina, N.; Larionov, V. TAR cloning: Insights into gene function, long-range haplotypes and genome structure and evolution. Nat. Rev. Genet. 2006, 7, 805–812. [Google Scholar] [CrossRef]
- Kouprina, N.; Larionov, V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008, 3, 371–377. [Google Scholar] [CrossRef]
- Kouprina, N.; Larionov, V. Transformation-associated recombination (TAR) cloning and its applications for gene function; genome architecture and evolution; biotechnology and biomedicine. Oncotarget 2023, 14, 1009–1033. [Google Scholar] [CrossRef]
- Noskov, V.N.; Kouprina, N.; Leem, S.H.; Ouspenski, I.; Barrett, J.C.; Larionov, V. A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genom. 2003, 4, 16. [Google Scholar] [CrossRef]
- Nikiforuk, A.M.; Leung, A.; Cook, B.W.M.; Court, D.A.; Kobasa, D.; Theriault, S.S. Rapid one-step construction of a Middle East Respiratory Syndrome (MERS-CoV) infectious clone system by homologous recombination. J. Virol. Methods 2016, 236, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Thi Nhu Thao, T.; Labroussaa, F.; Ebert, N.; V’Kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020, 582, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zhang, X.; Wu, H.; Liu, C.; Liu, J.; Hu, X.; Qu, L. Assessment of the IFN-β response to four feline caliciviruses: Infection in CRFK cells. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 34, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.C.; Scott, F.W. Antibody-mediated enhancement of disease in feline infectious peritonitis: Comparisons with dengue hemorrhagic fever. Comp. Immunol. Microbiol. Infect. Dis. 1981, 4, 175–189. [Google Scholar] [CrossRef]
- Giori, L.; Giordano, A.; Giudice, C.; Grieco, V.; Paltrinieri, S. Performances of different diagnostic tests for feline infectious peritonitis in challenging clinical cases. J. Small Anim. Pract. 2011, 52, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Tekes, G.; Spies, D.; Bank-Wolf, B.; Thiel, V.; Thiel, H.J. A reverse genetics approach to study feline infectious peritonitis. J. Virol. 2012, 86, 6994–6998. [Google Scholar] [CrossRef]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef]
- Bailey-Elkin, B.A.; Knaap, R.C.; Johnson, G.G.; Dalebout, T.J.; Ninaber, D.K.; van Kasteren, P.B.; Bredenbeek, P.J.; Snijder, E.J.; Kikkert, M.; Mark, B.L. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 2014, 289, 34667–34682. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Xia, C.; Kang, H.; Jia, H. The ORF1ab of Feline Coronavirus Plays a Critical Role in Regulating the Innate Immune Response. Viruses 2025, 17, 1282. https://doi.org/10.3390/v17091282
Gu H, Xia C, Kang H, Jia H. The ORF1ab of Feline Coronavirus Plays a Critical Role in Regulating the Innate Immune Response. Viruses. 2025; 17(9):1282. https://doi.org/10.3390/v17091282
Chicago/Turabian StyleGu, Haorong, Chuqiao Xia, Hongtao Kang, and Honglin Jia. 2025. "The ORF1ab of Feline Coronavirus Plays a Critical Role in Regulating the Innate Immune Response" Viruses 17, no. 9: 1282. https://doi.org/10.3390/v17091282
APA StyleGu, H., Xia, C., Kang, H., & Jia, H. (2025). The ORF1ab of Feline Coronavirus Plays a Critical Role in Regulating the Innate Immune Response. Viruses, 17(9), 1282. https://doi.org/10.3390/v17091282





