-
 Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism
Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism -
 Assessment of Organic Pollutants Desorbed from Plastic Litter Items Stranded on Cadiz Beaches (SW Spain)
Assessment of Organic Pollutants Desorbed from Plastic Litter Items Stranded on Cadiz Beaches (SW Spain) -
 Distribution of Legacy and Emerging PFAS in a Terrestrial Ecosystem Located near a Fluorochemical Manufacturing Facility
Distribution of Legacy and Emerging PFAS in a Terrestrial Ecosystem Located near a Fluorochemical Manufacturing Facility -
 Application of the AI-Based Framework for Analyzing the Dynamics of Persistent Organic Pollutants (POPs) in Human Breast Milk
Application of the AI-Based Framework for Analyzing the Dynamics of Persistent Organic Pollutants (POPs) in Human Breast Milk -
 Embryonic Exposure to TPhP Elicits Osteotoxicity via Metabolic Disruption in Oryzias latipes
Embryonic Exposure to TPhP Elicits Osteotoxicity via Metabolic Disruption in Oryzias latipes
Journal Description
Toxics
Toxics
is an international, peer-reviewed, open access journal on all aspects of the toxic chemicals and materials, published monthly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, Embase, CAPlus / SciFinder, AGRIS, and other databases.
- Journal Rank: JCR - Q1 (Toxicology) / CiteScore - Q1 (Chemical Health and Safety)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 18.1 days after submission; acceptance to publication is undertaken in 1.9 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
4.1 (2024);
5-Year Impact Factor:
4.6 (2024)
Latest Articles
A Systematic Review of Aircraft Disinsection Safety, Toxicity, and Tolerability
Toxics 2025, 13(11), 965; https://doi.org/10.3390/toxics13110965 (registering DOI) - 9 Nov 2025
Abstract
Treatment of aircraft with insecticide in a procedure referred to as ‘disinsection’ is recommended to prevent the conveyance of arthropod vectors internationally and to mitigate the globalization of vector-borne infectious diseases. However, the full spectrum of human-based outcomes related to disinsection of conveyances
[...] Read more.
Treatment of aircraft with insecticide in a procedure referred to as ‘disinsection’ is recommended to prevent the conveyance of arthropod vectors internationally and to mitigate the globalization of vector-borne infectious diseases. However, the full spectrum of human-based outcomes related to disinsection of conveyances has not been recently synthesized. A systematic review was conducted to evaluate the human safety and toxicity of insecticides used during the process of disinsecting international aircraft, marine vessels, rail, and ground transportation of mosquitoes. The systematic review was conducted according to the PRISMA guidelines and was registered in PROSPERO (CRD42024543998). The certainty of the evidence was rated, and key primary outcomes, including human health effects of conveyance disinsection, were synthesized. A total of 21 studies that described human health effects of conveyance disinsection and reported outcomes of safety, toxicity, and tolerability were included, and were of generally limited quality and high risk of bias, with low to very low certainty of estimates of effect. No high-quality studies investigating the safety, toxicity, or tolerability of disinsection were identified. Human health effects, including morbidity, including work days lost, adverse events including hospitalization, objective measures of insecticide toxicity, detectable and elevated urinary metabolites, and subjective reporting of symptoms consistent with acute insecticide poisoning, were reported by the small number of uncontrolled observational studies and public health surveillance reports included. Given the reports of significant morbidity, adverse events, and toxicity putatively attributable to aircraft disinsection, well-designed studies in exposed populations investigating the full range of human health impacts of disinsection on passengers and crew are urgently needed.
Full article
(This article belongs to the Special Issue Exposure to Chemicals and Health Effects in Occupational and Everyday Settings)
►
Show Figures
Open AccessFeature PaperReview
Dioxins and the One Health Paradigm: An Interdisciplinary Challenge in Environmental Toxicology
by
Marília Cristina Oliveira Souza and Jose L. Domingo
Toxics 2025, 13(11), 964; https://doi.org/10.3390/toxics13110964 - 7 Nov 2025
Abstract
Dioxins are legacy and persistent environmental pollutants that pose complex and far-reaching risks to human, animal, and ecosystem health. As unintentional byproducts of industrial and combustion processes, dioxins accumulate in the environment, biomagnify through food webs, and exert toxic effects even at low
[...] Read more.
Dioxins are legacy and persistent environmental pollutants that pose complex and far-reaching risks to human, animal, and ecosystem health. As unintentional byproducts of industrial and combustion processes, dioxins accumulate in the environment, biomagnify through food webs, and exert toxic effects even at low concentrations. This review applies a One Health lens to synthesize current knowledge on dioxin sources, environmental fate, exposure pathways, and toxicological impacts across species. We have critically examined existing surveillance systems, regulatory frameworks, and policy responses, highlighting both achievements and persistent gaps. A fully integrated One Health approach, linking environmental, animal, and human health domains, is essential for effective monitoring, risk assessment, and mitigation. It includes cross-sectoral collaboration, harmonized biomonitoring, evidence-based policy interventions, and transparent risk communication. Emerging evidence on climate-driven dioxin remobilization and microplastic interactions further underscores the urgency of adaptive, system-based strategies. Strengthening global capacity through such integrative approaches is vital to safeguard planetary health from these enduring contaminants. Quantitative insights and illustrative examples support these conclusions.
Full article
(This article belongs to the Section Toxicity Reduction and Environmental Remediation)
►▼
Show Figures

Figure 1
Open AccessArticle
Hidden Contamination Patterns: A Stochastic Approach to Assessing Unsymmetrical Dimethylhydrazine Transformation Products in Kazakhstan’s Rocket Crash Area
by
Ivan Radelyuk, Aray Zhakupbekova, Alua Zhumadildinova, Artem Kashtanov and Nassiba Baimatova
Toxics 2025, 13(11), 963; https://doi.org/10.3390/toxics13110963 - 6 Nov 2025
Abstract
Unsymmetrical dimethylhydrazine (UDMH), a highly toxic rocket propellant, remains a significant environmental concern in Kazakhstan due to repeated rocket stage falls near the Baikonur Cosmodrome. This study integrates chemical analysis with stochastic contamination transport modeling to evaluate the persistence and migration of UDMH
[...] Read more.
Unsymmetrical dimethylhydrazine (UDMH), a highly toxic rocket propellant, remains a significant environmental concern in Kazakhstan due to repeated rocket stage falls near the Baikonur Cosmodrome. This study integrates chemical analysis with stochastic contamination transport modeling to evaluate the persistence and migration of UDMH transformation products (TPs) in soils collected 15 years after the rocket crash. Vacuum-assisted headspace solid-phase microextraction coupled with gas chromatography–mass spectrometry (Vac-HS-SPME-GC-MS) was used to determine five major TPs. Among these, pyrazine (PAN) and 1-methyl-1H-pyrazole (MPA) were consistently detected at concentrations ranging from 0.04–2.35 ng g−1 and 0.06–3.48 ng g−1, respectively. Stochastic simulations performed with HYDRUS-1D indicated that the long-term persistence of these compounds is mainly controlled by physical nonequilibrium transport processes, including diffusion-limited exchange, weak sorption, and slow inter-domain mass transfer, rather than by degradation. Sensitivity analysis demonstrated that low dispersivity and diffusion coefficients enhance solute retention within immobile domains, maintaining residual levels over extended periods. The results demonstrate the efficacy of combined long-term monitoring and predictive modeling frameworks for assessing contamination dynamics in rocket impact zones.
Full article
(This article belongs to the Topic Water-Soil Pollution Control and Environmental Management)
►▼
Show Figures

Graphical abstract
Open AccessFeature PaperArticle
Preconception Hair Mercury and Serum Omega-3 Fatty Acids in Relation to Gestational Weight Gain Among Women Seeking Fertility Care
by
Han Han, Xinxiu Liang, Xilin Shen, Paige L. Williams, Tamarra James-Todd, Yazeed Allan, Roe P. Keshet, Jennifer B. Ford, Kathryn M. Rexrode, Jorge E. Chavarro, Russ Hauser and Lidia Mínguez-Alarcón
Toxics 2025, 13(11), 962; https://doi.org/10.3390/toxics13110962 - 6 Nov 2025
Abstract
Few studies have prospectively examined the joint effect of mercury (Hg) and omega-3 long-chain polyunsaturated fatty acids on gestational weight gain (GWG). This exploratory study included 120 women from the Environment and Reproductive Health (EARTH) study with preconception measurements of hair Hg and
[...] Read more.
Few studies have prospectively examined the joint effect of mercury (Hg) and omega-3 long-chain polyunsaturated fatty acids on gestational weight gain (GWG). This exploratory study included 120 women from the Environment and Reproductive Health (EARTH) study with preconception measurements of hair Hg and serum eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), followed through pregnancy. Linear regression was used to examine associations between hair Hg and GWG, while logistic regression estimated the odds of inadequate GWG based on National Academy of Medicine recommendations. In unadjusted models, higher hair Hg (≥1 vs. <1 ppm) was associated with lower GWG (β = −1.89; 95% CI: −3.70, −0.08) and increased odds of insufficient GWG (OR = 2.27; 95% CI: 1.00, 5.18). However, after multivariable adjustment including serum EPA + DHA, these associations were attenuated and became non-significant. A negative, though not statistically significant, association between hair Hg and GWG was observed among women in the lowest tertile of serum EPA + DHA (≥1 vs. <1 ppm: β = −3.26; 95% CI: −7.69, 1.17), whereas no such association was observed among those in higher tertiles (β [95% CI] = 0.44 [−4.21, 5.09] and −1.05 [−4.13, 2.02], respectively). Our findings suggest that the association between preconception Hg exposure and insufficient GWG may differ by serum EPA + DHA, but results require confirmation in cohorts with larger sample sizes.
Full article
(This article belongs to the Special Issue Environmental and Reproductive Epidemiology: Focus on Air Pollution and Associated Contaminants)
►▼
Show Figures

Figure 1
Open AccessArticle
Assessment of Heavy Metal Pollution in Mangrove Sediments of Liusha Bay, Leizhou Peninsula, China
by
Xianhui Yang, Huamei Huang, Ping Hu, Hong Luan, Bei Song, Zhaoyong Zheng, Cuiping Zhang, Ran Yan and Kang Li
Toxics 2025, 13(11), 961; https://doi.org/10.3390/toxics13110961 - 6 Nov 2025
Abstract
Heavy metal pollution threatens coastal ecosystems. Mangrove sediments, as transitional zones, are prone to contaminant accumulation. This study investigated eight heavy metals (Cu, Pb, Ni, As, Cr, Zn, Cd, Co) in Liusha Bay (Leizhou Peninsula, China). Field sampling, lab analysis, and multivariate statistics
[...] Read more.
Heavy metal pollution threatens coastal ecosystems. Mangrove sediments, as transitional zones, are prone to contaminant accumulation. This study investigated eight heavy metals (Cu, Pb, Ni, As, Cr, Zn, Cd, Co) in Liusha Bay (Leizhou Peninsula, China). Field sampling, lab analysis, and multivariate statistics were used to assess pollution sources and ecological risks. The results show Al and Fe dominate sediment composition, with elevated P, Mn, and Sr. Arsenic (As) exhibiting the highest pollution severity (50% sites moderately contaminated by Igeo). Enrichment factors (EF) indicate anthropogenic contributions to As, Cu, Ni, and Co, while Cd and Pb originate mainly from natural sources. Ecological risk assessments highlight moderate risks for As and Cd at some sites. Source analysis identifies three dominant pathways: (1) lithogenic inputs (volcanic rock weathering) contributing Fe, Zn, Cr, and Ni; (2) biogenic materials (calcium carbonate-secreting organisms) influencing Cu, Mn, and Cd; and (3) anthropogenic activities (aquaculture, maritime traffic) linked to Cu and Pb. This study emphasizes localized monitoring of As and Cd in mangroves and calls for the integrated management of natural and anthropogenic drivers to mitigate pollution risks.
Full article
(This article belongs to the Section Ecotoxicology)
►▼
Show Figures

Figure 1
Open AccessArticle
Analysis of the Prevalence of Alcohol and Psychoactive Substances Among Drivers in the Material from the Department of Forensic Medicine at the Medical University of Bialystok in Poland
by
Michal Szeremeta, Julia Janica, Gabriela Jurkiewicz, Marta Galicka, Julia Koścień, Julia Więcko, Jakub Perkowski, Michal Krzysztof Jeleniewski, Karol Siemieniuk and Anna Niemcunowicz-Janica
Toxics 2025, 13(11), 960; https://doi.org/10.3390/toxics13110960 - 6 Nov 2025
Abstract
In recent years, the issue of drivers under the influence of medications and psychoactive substances as a cause of road accidents has gained increasing importance. This study aimed to assess the prevalence and blood concentration ranges of alcohol and psychoactive substances among drivers
[...] Read more.
In recent years, the issue of drivers under the influence of medications and psychoactive substances as a cause of road accidents has gained increasing importance. This study aimed to assess the prevalence and blood concentration ranges of alcohol and psychoactive substances among drivers in northeastern Poland between 2013 and 2024. To determine the prevalence of medications and psychoactive substances in drivers’ blood, data were collected from 266 blood samples obtained from drivers (251 men and 15 women). Among these, 79 drivers died immediately, 61 drivers survived the accident, and 126 drivers were stopped for roadside checks. The presence of the studied substances was confirmed using gas chromatography combined with mass spectrometry detection (GC-MS) and liquid chromatography combined with mass spectrometry detection (LC-MS). Blood alcohol content was measured using headspace gas chromatography with a flame ionisation detector (HS-GC-FID). Psychoactive substances were detected in 152 of the 266 samples. Drivers testing positive for medications and psychoactive substances were most frequently stopped during roadside controls—67.46%. Among the total positive cases, psychoactive substances used alone or in combination included THC—46.3% (range 0.2–20 ng/mL), alcohol—26.8% (range 0.1–4.1‰), amphetamines—20.7% (range 15–2997 ng/mL), opiates—4.3% (morphine 66.0 ng/mL; methadone 174.0 ng/mL; ranges: tramadol 15.0–600.0 ng/mL; fentanyl 45.0–100.0 ng/mL), benzodiazepines—9.8% (ranges: diazepam 55.0–480.0 ng/mL; midazolam 17.0–1200.0 ng/mL; clonazepam 21.0–36.0 ng/mL), stimulants—6.10% (ranges: amphetamine 15.0–2997.0 ng/mL; cocaine 4.0–30.0 ng/mL; benzoylecgonine 38.0–602.0 ng/mL; PMMA 45.0–360.0 ng/mL; MDMA 20.0–75.0 ng/mL; mephedrone 37.5 ng/mL; alfa-PVP 120 ng/mL), psychotropic drugs—3.1% (carbamazepine 8.0–2100.0 ng/mL; zolpidem 233.0 ng/mL; citalopram 320.0 ng/mL; opipramol 220 ng/mL). The most commonly used substance among car and motorcycle drivers was THC (37.7% of car drivers and 60% of motorcyclists). Among operators of other types of vehicles, alcohol was the most frequently detected substance, present in 35% of cases. The majority of drivers (81.1%) were under the influence of a single substance. Among the drivers, 7.3% consumed alcohol in combination with at least one other substance, and 11.6% used two or more substances excluding alcohol. Among the psychoactive substances most frequently used alone or in combination with others, THC was predominant. Roadside testing, based on effects similar to alcohol intoxication, was mainly conducted on male drivers.
Full article
(This article belongs to the Special Issue Current Issues and Research Perspectives in Forensic Toxicology)
►▼
Show Figures

Graphical abstract
Open AccessArticle
Bioremediation of High-Concentration Heavy Metal-Contaminated Soil by Combined Use of Acidithiobacillus ferrooxidans and Fe3O4–GO Anodes
by
Alifeila Yilahamu, Xuewen Wu, Xiaonuan Wang, Shengjuan Peng and Weihua Gu
Toxics 2025, 13(11), 959; https://doi.org/10.3390/toxics13110959 - 6 Nov 2025
Abstract
Soils heavily contaminated with potentially toxic elements (PTEs) pose substantial risks to the environment and human health. However, conventional remediation methods are often plagued by high energy consumption and the potential for secondary pollution. To address this challenge, this study developed a synergistic
[...] Read more.
Soils heavily contaminated with potentially toxic elements (PTEs) pose substantial risks to the environment and human health. However, conventional remediation methods are often plagued by high energy consumption and the potential for secondary pollution. To address this challenge, this study developed a synergistic system combining acidophilic bacteria with a Fe-modified anode, aiming to enhance the remediation of PTEs in such contaminated soils. This system integrates the following three core components: the catalytic function of Fe3O4–graphene-oxide (Fe3O4–GO) nanocomposites, the acclimation of microbial communities, and the optimization of process parameters—specifically, applied electric current, pH, and oxidation–reduction potential (ORP). Experimental treatments were designed to assess the individual and combined effects of three key factors: bacterial inoculation, the Fe-modified anode, and the addition of Fe3O4–GO. The results revealed that the integrated synergistic system effectively reduced the soil pH from 2.9 to 2.0 and maintained the ORP at approximately 600 mV. For PTE removal, the system achieved efficiencies of 89% for Zn, 85.89% for Cu, 66.3% for Pb, 77.89% for Cd, and 40.63% for Cr, respectively. In contrast, control groups lacking bacteria, applied current, or Fe3O4–GO exhibited significantly lower metal removal efficiencies. Notably, the bacteria-free treatment led to a more than 50% reduction in Cr removal. Additionally, the group with an unmodified anode only achieved 1/3 to 1/2 of the removal efficiencies observed in the full synergistic system; this discrepancy is likely attributed to reduced electron transfer efficiency and compromised microbial adhesion on the anode surface. These findings demonstrate that the coupling of electrochemical enhancement, acidophilic microbial activity, and Fe3O4–GO catalysis constitutes an effective and energy-efficient approach for remediating soils contaminated with high concentrations of PTEs while simultaneously minimizing the risk of secondary pollution.
Full article
(This article belongs to the Section Toxicity Reduction and Environmental Remediation)
►▼
Show Figures

Figure 1
Open AccessArticle
Association Between Pyrethroid Exposure Levels and Obesity/Cardiovascular Indicators in Korean Adults: Focused on the 2nd National Environmental Health Survey (2012–2014)
by
Eunbee Bang, Youngwook Lim and Kiyoun Kim
Toxics 2025, 13(11), 958; https://doi.org/10.3390/toxics13110958 - 5 Nov 2025
Abstract
Background/Objectives: This study evaluated associations between urinary 3-phenoxybenzoic acid (3-PBA), a metabolite of pyrethroids, and cardiometabolic indicators in a nationally representative sample of Korean adults using data from the 2nd Korean National Environmental Health Survey (KoNEHS 2012–2014). Methods: Urinary 3-PBA concentrations
[...] Read more.
Background/Objectives: This study evaluated associations between urinary 3-phenoxybenzoic acid (3-PBA), a metabolite of pyrethroids, and cardiometabolic indicators in a nationally representative sample of Korean adults using data from the 2nd Korean National Environmental Health Survey (KoNEHS 2012–2014). Methods: Urinary 3-PBA concentrations were creatinine-adjusted; participants with urinary creatinine < 0.3 or > 3.0 g/L were excluded. Associations with triglycerides, BMI, HDL cholesterol, TSH, and T4 were analyzed using non-parametric tests and multiple regression, with additional verification through log-transformed variables and multiple-comparison control. Results: Urinary 3-PBA levels were higher in females, increased with age, and were elevated among rural residents and frequent pesticide users. Triglycerides and TSH showed positive associations with 3-PBA, whereas T4 showed a negative association. BMI displayed a weak negative correlation without consistent significance, and HDL cholesterol was not statistically significant. In multiple regression models, triglycerides, TSH, and T4 remained significantly associated with urinary 3-PBA. Conclusions: Statistically significant associations were observed between urinary 3-PBA concentrations and several cardiometabolic indicators, including triglycerides, TSH, and T4, in Korean adults. These findings suggest that even low-level environmental exposure to pyrethroids may influence lipid metabolism and thyroid function. Given the cross-sectional design and the short biological half-life of 3-PBA, the results should be interpreted as associations rather than causation, highlighting the need for longitudinal studies and continued biomonitoring.
Full article
(This article belongs to the Section Human Toxicology and Epidemiology)
Open AccessArticle
Airborne Dental Material Particulates and Occupational Exposure: Computational and Field Insights into Airflow Dynamics and Control Strategies
by
Chanapat Chanbandit, Kanchana Kanchanatawewat, Ghaim Man Oo, Jatuporn Thongsri and Kuson Tuntiwong
Toxics 2025, 13(11), 957; https://doi.org/10.3390/toxics13110957 - 5 Nov 2025
Abstract
Occupational exposure to airborne polymethacrylate (PMMA) particles during dental laboratory procedures poses an underexplored health risk. This study presents the first integrated Computational Fluid Dynamics (CFD) and real-time particle monitoring investigation of 0.5 µm PMMA particle dispersion during mechanical polishing in an actual
[...] Read more.
Occupational exposure to airborne polymethacrylate (PMMA) particles during dental laboratory procedures poses an underexplored health risk. This study presents the first integrated Computational Fluid Dynamics (CFD) and real-time particle monitoring investigation of 0.5 µm PMMA particle dispersion during mechanical polishing in an actual clinic. We quantitatively assessed particle behavior in 30 s exposure scenarios by examining the effects of dental professional work orientations and comparing two mitigation strategies, rear-inlet portable air cleaners (PACs) and a Box Dust Collector (BC), with an emphasis on the safety of both personnel and patients. The findings establish that operatory airflow is a primary safety determinant: aligning the workflow with the main airflow (0°). Furthermore, the combined use of PACs and BC demonstrated synergistic superiority, achieving the optimal reduction in peak concentrations and airborne residence time. PACs alone reduced working zone concentrations by up to 80%, while BC provided a crucial 40–60 s delay in initial plume dispersion. We conclude that effective exposure control requires a proactive, two-stage engineering defense: source confinement augmented by continuous ambient filtration. This research provides a robust, evidence-based foundation for defining airflow-aware ergonomic and combined engineering standards in the evolving digital era of dentistry.
Full article
(This article belongs to the Special Issue Exposure to Chemicals and Health Effects in Occupational and Everyday Settings)
►▼
Show Figures

Graphical abstract
Open AccessArticle
From Soil to Plate: Lithium and Other Trace Metals Uptake in Vegetables Under Variable Soil Conditions
by
Nadia Paun, Ramona Zgavarogea, Violeta-Carolina Niculescu, Ana Maria Nasture, Iulian Voicea and Diana Ionela Popescu (Stegarus)
Toxics 2025, 13(11), 956; https://doi.org/10.3390/toxics13110956 - 5 Nov 2025
Abstract
The bioaccumulation of trace metals in edible crops is a key pathway of dietary exposure, with direct implications for environmental health and food safety. This study specifically investigated the bioaccumulation and soil–plant transfer of lithium (Li) in edible crops, alongside other selected trace
[...] Read more.
The bioaccumulation of trace metals in edible crops is a key pathway of dietary exposure, with direct implications for environmental health and food safety. This study specifically investigated the bioaccumulation and soil–plant transfer of lithium (Li) in edible crops, alongside other selected trace metals (Cu, Mn, Sr, Zn), to understand its unique environmental mobility and dietary exposure risks in onion, garlic, green salad, cucumber, and zucchini cultivated across Romania. Forty-two paired samples of vegetable tissues and rhizosphere soils were collected from eleven agricultural regions, and were analyzed using spectroscopic techniques. Soils were predominantly neutral to slightly acidic, conditions that significantly affected metal mobility and uptake. Results revealed element-specific decoupling between soil and plant concentrations. Essential micronutrients (Zn, Cu) showed higher transfer factors, consistent with active physiological uptake, while toxic non-essential metals (Pb, Cd) remained below European regulatory thresholds, reflecting effective exclusion mechanisms. Lithium exhibited spatially heterogeneous transfer patterns, strongly influenced by local geochemical variability. Curvilinear soil–plant relationships for Fe, Zn, Sr, Mn, Cu, and Li further underscored the role of soil chemistry in shaping translocation. These findings establish a robust baseline for assessing dietary risks, confirming the current low-risk status of vegetables in the surveyed regions, and provide valuable guidance for sustainable agricultural management and food safety monitoring.
Full article
(This article belongs to the Special Issue Exploring Toxicity, Metabolism, and Transformation of Organic Pollutants)
►▼
Show Figures

Figure 1
Open AccessArticle
Preparation of Perovskite-Type LaCoO3 and Its Catalytic Degradation of Formaldehyde in Wastewater
by
Qingguo Ma, Qin Gao, Shancheng Li, Tianying Li, Zhiqian Fan, Binglong Mu and Yike Zhang
Toxics 2025, 13(11), 955; https://doi.org/10.3390/toxics13110955 - 5 Nov 2025
Abstract
Removing toxic formaldehyde (HCHO) from environmental water is crucial for human health and the ecosystem. Perovskite-type Lanthanum cobalt oxide (LaCoO3) has achieved great success in a wide range of catalytic processes; however, this concept has been rarely applied to the degradation
[...] Read more.
Removing toxic formaldehyde (HCHO) from environmental water is crucial for human health and the ecosystem. Perovskite-type Lanthanum cobalt oxide (LaCoO3) has achieved great success in a wide range of catalytic processes; however, this concept has been rarely applied to the degradation of HCHO. Here, we prepared perovskite-type catalysts with different La/Co molar ratios, and the time for HCHO oxidation degradation at room temperature was shortened by 12 times (10 min vs. 119 min) compared to other heterogeneous catalysts. LaCoO3 exhibits superior catalytic activity for HCHO degradation at room temperature when the La/Co molar ratio is 1:1 compared to lanthanum cobalt oxides with other molar ratios. The X-ray photoelectron spectroscopy (XPS) test results show that increasing the La/Co molar ratio reduces the Co2+ content in the catalyst, while Co2+ plays the most important role in the catalyst. Quencher experiments indicated that sulfate radicals (SO4·−) and hydroxyl radicals (·OH) were the primary reactive species for the removal of HCHO. This finding suggests that the catalytic oxidation reaction involving HCHO operates as a heterogeneous Fenton-like oxidation reaction.
Full article
(This article belongs to the Special Issue Toxic Agent Analysis and Removal for Healthy Living and Safe Environment)
►▼
Show Figures

Graphical abstract
Open AccessArticle
Effects of Prenatal Essential and Toxic Metal Exposure on Children’s Neurodevelopment: A Multi-Method Approach
by
Xiruo Kou, Stefano Renzetti, Josefa Canals, Stefano Calza, Cristina Jardí and Victoria Arija
Toxics 2025, 13(11), 954; https://doi.org/10.3390/toxics13110954 - 5 Nov 2025
Abstract
The impact of prenatal exposure to trace metal mixtures on children’s neurodevelopment remains debated. Many studies treat all trace metals as a single entity, overlooking the distinct biological roles of essential and toxic metals. This approach may highlight overall exposure but fails to
[...] Read more.
The impact of prenatal exposure to trace metal mixtures on children’s neurodevelopment remains debated. Many studies treat all trace metals as a single entity, overlooking the distinct biological roles of essential and toxic metals. This approach may highlight overall exposure but fails to capture their differential effects on neurodevelopment. This study aims to examine the associations between prenatal exposure to essential and toxic metals and children’s cognitive development, focusing on their independent effects. A cohort of 201 mother–infant pairs was analyzed. Maternal urinary metal levels were measured at 12 weeks of gestation, and children’s neurodevelopment was assessed at 4 years using the Wechsler Preschool and Primary Scale of Intelligence and the Developmental Neuropsychological Assessment. Generalized Additive Models (GAM), Restricted Cubic Spline (RCS), and Weighted Quantile Sum (WQS) regression were applied. GAM identified non-linear associations between essential metals (manganese and molybdenum) and cognitive outcomes, including verbal comprehension index (VCI), working memory index, full-scale IQ, and general ability index, which were confirmed by RCS. No non-linear relationships were observed for toxic metals. WQS showed negative associations between toxic metals and VCI (b = −1.07), processing speed index (b = −0.98), vocabulary acquisition index (b = −1.25), and verbal fluency (b = −0.23), mainly driven by cadmium (Cd) and antimony (Sb). Essential metal mixtures were not associated with cognitive outcomes. Prenatal exposure to toxic metals negatively affects children’s cognitive and neuropsychological development. Reducing maternal exposure during pregnancy is essential for protecting offspring development.
Full article
(This article belongs to the Special Issue Toxicity and Safety Assessment of Exposure to Heavy Metals)
►▼
Show Figures

Figure 1
Open AccessArticle
Graph Neural Network-Based Toxicity Prediction by Integrating Molecular Fingerprints and Knowledge Graph Features
by
Junjie Xie, Wei Liu, Wei Hu, Mei Ouyang and Tingting Huang
Toxics 2025, 13(11), 953; https://doi.org/10.3390/toxics13110953 - 5 Nov 2025
Abstract
Molecular toxicity prediction plays a crucial role in drug screening and environmental health risk assessment. Traditional toxicity prediction models primarily rely on molecular fingerprints and other structural features, while neglecting the complex biological mechanisms underlying compound toxicity, resulting in limited predictive accuracy, poor
[...] Read more.
Molecular toxicity prediction plays a crucial role in drug screening and environmental health risk assessment. Traditional toxicity prediction models primarily rely on molecular fingerprints and other structural features, while neglecting the complex biological mechanisms underlying compound toxicity, resulting in limited predictive accuracy, poor interpretability, and reduced generalizability. To address this challenge, this study proposes a novel molecular toxicity prediction framework that integrates knowledge graphs with Graph Neural Networks (GNNs). Specifically, we constructed a heterogeneous toxicological knowledge graph (ToxKG) based on ComptoxAI. ToxKG incorporates data from authoritative databases such as PubChem, Reactome, and ChEMBL, and covers multiple entities and relationships including chemicals, genes, signaling pathways, and bioassays. We then systematically evaluated six representative GNN models (GCN, GAT, R-GCN, HRAN, HGT, and GPS) on the Tox21 dataset. Experimental results demonstrate that heterogeneous graph models enriched with ToxKG information significantly outperform traditional models relying solely on structural features across multiple metrics including AUC, F1-score, ACC, and balanced accuracy (BAC). Notably, the GPS model achieved the highest AUC value (0.956) for key receptor tasks such as NR-AR, highlighting the critical role of biological mechanism information and heterogeneous graph structures in toxicity prediction. This study provides a promising pathway toward the development of interpretable and efficient intelligent models for toxicological risk assessment.
Full article
(This article belongs to the Section Novel Methods in Toxicology Research)
►▼
Show Figures

Graphical abstract
Open AccessArticle
Source-Oriented Health Risk Assessment of Potentially Toxic Elements in the Water-Soil-Crop System Using Monte Carlo Simulation: A Case Study of the Laoguan River Basin, China
by
Xiaolin Jia, Hui Fu, Ding Ding, Xi Ren, Pei Zhao, Xidong Chen, Xiaonan Luo, Baojian Guo, Hongbin Xu, Zhiwei Sheng and Haitao Huang
Toxics 2025, 13(11), 952; https://doi.org/10.3390/toxics13110952 - 4 Nov 2025
Abstract
Mining and smelting release potentially toxic elements (PTEs) that threaten ecosystems and public health. However, comprehensive risk assessments of PTEs across environmental media near mining areas remain scarce. The Laoguan River Basin is located in southwestern Henan Province, China. It lies within the
[...] Read more.
Mining and smelting release potentially toxic elements (PTEs) that threaten ecosystems and public health. However, comprehensive risk assessments of PTEs across environmental media near mining areas remain scarce. The Laoguan River Basin is located in southwestern Henan Province, China. It lies within the water source area of China’s South-to-North Water Diversion Middle Route Project. This area has high geographic and ecological importance. In this study, we analyzed the pollution characteristics of PTEs in the water–soil–crop system. We also performed a source-oriented health risk assessment by integrating Monte Carlo simulation with source apportionment. According to this study, Mo and Sb were the predominant contaminants in soils and water. Pb, Cr, and Ni were elevated in crops. The health risk assessment indicated that PTEs in surface water were at acceptable levels. In contrast, PTEs in soils pose both non-carcinogenic and carcinogenic risks, particularly to children. The estimated risks were 1% (non-carcinogenic) and 64% (carcinogenic), with ingestion as the primary exposure pathway. Source apportionment showed that the surface water pollution was mainly linked to diverse mining activities. Soil pollution was jointly influenced by the geological background and mining and agricultural activities. Crop pollution was primarily associated with mining and agricultural activities. Geological background and mining were the main driving factors of the increased health risks for children. They accounted for 83% of the non-carcinogenic risk and 79% of the carcinogenic risk. Overall, these results are crucial for pollution control, safeguarding public health and safety, and promoting balanced economic and ecological development.
Full article
(This article belongs to the Special Issue Pollutants in Soil and Groundwater, Effects and Strategies for Comprehensive Risk Assessment)
►▼
Show Figures

Figure 1
Open AccessArticle
Adsorption of Arsenic by Magnetically Modified Biochar from Mulberry Tree Stems
by
Sheng Hu, Jiayu Yang, Jingnan Zhang, Jing Pan, Liling Yan, Kun Dong, Dunqiu Wang, Ying Song and Meina Liang
Toxics 2025, 13(11), 951; https://doi.org/10.3390/toxics13110951 - 4 Nov 2025
Abstract
In this study, mulberry tree stems were used as raw material to prepare magnetic modified biochar, Fe-BC-500, using the co-precipitation method. The structure of Fe-BC-500 was systematically characterized and tested for arsenic (As) adsorption in batch experiments by varying parameters such as solution
[...] Read more.
In this study, mulberry tree stems were used as raw material to prepare magnetic modified biochar, Fe-BC-500, using the co-precipitation method. The structure of Fe-BC-500 was systematically characterized and tested for arsenic (As) adsorption in batch experiments by varying parameters such as solution pH (3–11), the concentrations of co-existing anions (2–20 mg/L), and ionic strength (0–0.5 mol/L NaNO3). The results indicate that Fe-BC-500 exhibited optimal adsorption capacity at pH 4 and an initial As(V) concentration of 20 mg/L. The influence of co-existing anions on As(V) adsorption followed the order PO43− > SO42− > NO3−. Kinetic analysis showed that adsorption of Fe-BC-500 on As(V) followed a pseudo-second-order kinetic model, with a correlation coefficient of 1.00, indicating chemical adsorption. The Langmuir model accurately described the isothermal adsorption results, indicating monolayer adsorption. Mechanistic analysis showed that As(V) was fixed on the Fe-BC-500 surface through complexation reactions, demonstrating adsorption specificity. This study provides a theoretical basis and highlights the application potential of magnetically modified biochar for removing As(V) from water.
Full article
(This article belongs to the Topic The Challenges and Future Trends in Anthropogenic and Natural Pollution Control Engineering)
►▼
Show Figures

Graphical abstract
Open AccessFeature PaperReview
Internal Exposure Levels and Health Risk Assessment of Melamine and Organophosphate Metabolites in Urine: Research Progress and Prospects
by
Qu Zhang, Qi Jiang, Xin-Hong Wang, Liang Wang, Mei-Hua Tian, Da-Zhong Chen, Chun-Yan Huo and Wen-Long Li
Toxics 2025, 13(11), 950; https://doi.org/10.3390/toxics13110950 - 4 Nov 2025
Abstract
►▼
Show Figures
With the widespread use of emerging contaminants such as melamine (MEL) and organophosphate esters (OPEs) as alternatives to traditional flame retardants, their ubiquitous presence in the environment has raised concerns about human internal exposure and health risks. Urine, as a critical matrix for
[...] Read more.
With the widespread use of emerging contaminants such as melamine (MEL) and organophosphate esters (OPEs) as alternatives to traditional flame retardants, their ubiquitous presence in the environment has raised concerns about human internal exposure and health risks. Urine, as a critical matrix for biomonitoring, enables accurate assessment of internal exposure to these contaminants and their metabolites. This review systematically summarizes the research progress on urinary biomonitoring of MEL and its derivatives (cyanuric acid (CYA), ammeline (AMN), ammelide (AMD)) and OPE metabolites. It covers analytical methods (sample pretreatment including enzymatic hydrolysis and extraction, instrumental detection via HPLC-MS/MS/UPLC-MS/MS, and method validation), exposure characteristics (global spatial differences, population disparities among sensitive groups like children and e-waste workers, and temporal variations such as postprandial peaks), and health risk assessments. Results show that MEL and CYA are widely detected in urine (detection rates > 97%), with CYA dominating total MEL (66.2–80%); OPE metabolites exhibit regional compositional differences, e.g., bis(2-chloroethyl) phosphate (BCEP) in Shenzhen and diphenyl phosphate (DPHP) in New York. Current exposure levels are generally safe, but 2–12% of sensitive individuals face potential risks. This review highlights key challenges (method standardization, limited hydroxylated OPE standards) and provides directions for future research to establish a comprehensive exposure–health risk evaluation system.
Full article
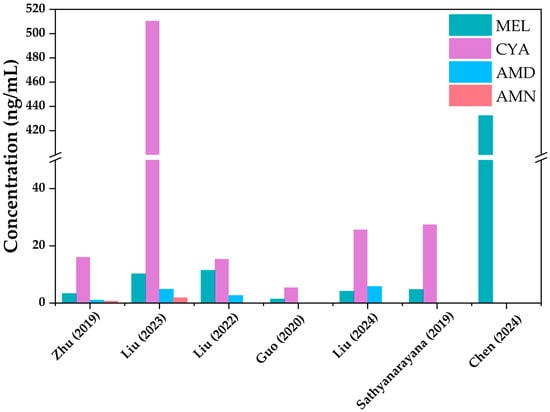
Figure 1
Open AccessReview
Microbial Degradation of Herbicide Residues in Australian Soil: An Overview of Mechanistic Insights and Recent Advancements
by
Imtiaz Faruk Chowdhury, Gregory S. Doran, Benjamin J. Stodart, Chengrong Chen and Hanwen Wu
Toxics 2025, 13(11), 949; https://doi.org/10.3390/toxics13110949 - 3 Nov 2025
Abstract
Herbicides are chemical compounds that are toxic to weed plants. Modern agriculture relies heavily on herbicides for the control of weeds to maximize crop yields. Herbicide usage in the Australian grains industry is estimated to have increased by more than 65% from 2014
[...] Read more.
Herbicides are chemical compounds that are toxic to weed plants. Modern agriculture relies heavily on herbicides for the control of weeds to maximize crop yields. Herbicide usage in the Australian grains industry is estimated to have increased by more than 65% from 2014 to 2024, which equates to more than AUD 2.50 billion dollars per year. The increased popularity of herbicides in farming systems has raised concerns about their negative impacts on the environment, human health and agricultural sustainability due to the rapid evolution of herbicide resistance, as well as their behaviour and fate in the soil. Due to excessive use of herbicides, soil and water pollution, reduced biodiversity and depression in soil heterotrophic bacteria (including denitrifying bacteria) and fungi are becoming increasingly common. Biological degradation governed by microorganisms serves as a major natural remediation process for a variety of pollutants including herbicides. This review provides a brief overview of the present status of herbicide residues in Australian farming systems, with a focus on the microbial degradation of herbicides in soil. It highlights key bacterial and fungal strains involved and the environmental factors influencing the biodegradation process. Recent advancements, including the application of omics technologies, are outlined to provide a comprehensive understanding of the biodegradation process.
Full article
(This article belongs to the Special Issue The Toxicity of Heavy Metals and Chemical Pollutants in Agricultural Soil and Plants: Ecological Risks and Remediation)
►▼
Show Figures

Figure 1
Open AccessArticle
From Safety Evaluation to Influencing Factors Analysis: A Comprehensive Investigation on Ocular Irritation of Baby Bath Products
by
Qidi He, Yurong Zhong, Peining Li, Yanhua Guo, Chengkai Mei, Dongmei Xu, Erping Yan, Shaofeng Xi, Guoshan He and Jianhua Tan
Toxics 2025, 13(11), 948; https://doi.org/10.3390/toxics13110948 - 3 Nov 2025
Abstract
Given the underdevelopment and sensitivity of babies’ eyes, frequently used baby bath products have garnered attention for their ocular irritation safety. The selection and application of raw materials are key factors. We tested 39 commercial baby bath products for ocular irritation safety and
[...] Read more.
Given the underdevelopment and sensitivity of babies’ eyes, frequently used baby bath products have garnered attention for their ocular irritation safety. The selection and application of raw materials are key factors. We tested 39 commercial baby bath products for ocular irritation safety and proposed a novel evaluation method using the “Risk Attention Index” to analyze formulation risk factors. The ocular irritation potential of 39 commonly used raw materials under typical usage conditions was investigated via animal and cell models. Results showed over 20% of commercial products had safety risks. Surfactants like cocamidopropyl betaine (CAPB), sodium laureth sulfate (SLES), and sodium lauroamphoacetate (SLA) had the highest “Risk Attention Index”, requiring special attention. Some fragrance components were also potential irritants. Combining SLES with panthenol, maltooligosyl glucoside/hydrogenated starch hydrolysate, and Tween®-28 could significantly reduce its irritation in an animal model. The same ingredients from different sources led to inconsistent irritation results. This suggests that manufacturing processes of raw materials, in addition to their chemical properties, concentration, and combination methods, all influence the ocular irritation potential. This study provides key scientific evidence for assessing ocular irritation and developing low-irritation formulations in baby bath products, thereby enhancing product safety.
Full article
(This article belongs to the Section Exposome Analysis and Risk Assessment)
►▼
Show Figures
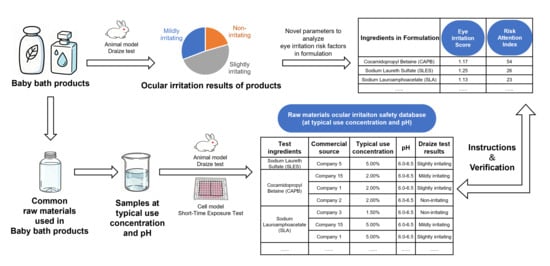
Graphical abstract
Open AccessReview
Nephrotoxicity of Phthalates: A Review Based on Epidemiological and Toxicological Evidence
by
Yuehang Wei, Minghui Zhang, Jiayuan Song, Tianyue Wang, Yuqin Ma, Liqiang Qin, Jiafu Li, Xiaoyan Qian and Jingsi Chen
Toxics 2025, 13(11), 947; https://doi.org/10.3390/toxics13110947 - 3 Nov 2025
Abstract
Phthalates are a widely used class of plasticizers known to cause various health issues. Although numerous review articles have addressed the multi-organ toxicities of Phthalates and their metabolites, a specialized review focusing on their nephrotoxicity remains scarce. In this study, the nephrotoxicity of
[...] Read more.
Phthalates are a widely used class of plasticizers known to cause various health issues. Although numerous review articles have addressed the multi-organ toxicities of Phthalates and their metabolites, a specialized review focusing on their nephrotoxicity remains scarce. In this study, the nephrotoxicity of Phthalates and their metabolites is summarized from the views of epidemiological and toxicological evidence. Epidemiological studies have demonstrated a correlation between Phthalate exposure and abnormal urinary albumin-to-creatinine ratio (ACR) as well as glomerular filtration rate (eGFR) in children. In contrast, for adults, the epidemiological evidence for the association between Phthalates and ACR/eGFR remains controversial, necessitating further investigation. In this review, we explore the potential mechanisms by which Phthalates and their metabolites may induce nephrotoxicity. These mechanisms include the following: (1) induction of oxidative stress in renal cells; (2) reduction in aldosterone levels; (3) dysregulation of the renin-angiotensin system; (4) activation of endoplasmic reticulum (ER) stress; (5) renal fibrosis; (6) disruption of sodium and water homeostasis; and (7) activation of the heat shock response defense system. Finally, based on the current understanding, we propose future research directions and necessary efforts to advance knowledge in this field.
Full article
(This article belongs to the Section Human Toxicology and Epidemiology)
►▼
Show Figures
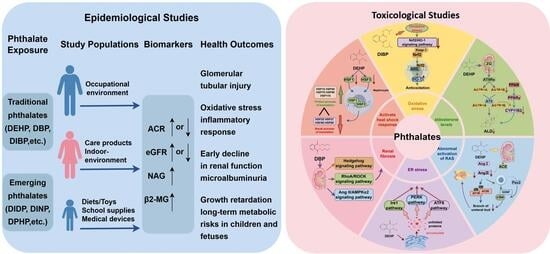
Graphical abstract
Open AccessArticle
Evaluating the Single and Combined Effects of BMDM and PS Microplastics on Chlorella sp.: Physiological and Transcriptomic Insights
by
Jin Liu, Yankun Zhang, Fengyuan Chen, Dandan Duan and Xiaoping Diao
Toxics 2025, 13(11), 946; https://doi.org/10.3390/toxics13110946 - 3 Nov 2025
Abstract
In the environment, the coexistence of microplastics (MPs) with other pollutants may either enhance or reduce the toxicity of MPs themselves or the co-occurring pollutants toward microalgae. This phenomenon is particularly notable when MPs interact with emerging pollutants, such as ultraviolet absorbers. This
[...] Read more.
In the environment, the coexistence of microplastics (MPs) with other pollutants may either enhance or reduce the toxicity of MPs themselves or the co-occurring pollutants toward microalgae. This phenomenon is particularly notable when MPs interact with emerging pollutants, such as ultraviolet absorbers. This study investigates the single and combined exposure effects of ultraviolet absorber (Butyl methoxydibenzoylmethane, BMDM, 50 μg/L) and MPs (Polystyrene, PS, 10 mg/L, d = 1 μm) on Chlorella sp. with a stress duration of 7 days. The results showed that cell density, chlorophyll a (Chla) concentration, and physical properties of cell surface integrity were higher in the combined stress group compared to the BMDM single stress group. Furthermore, transcriptome sequencing analysis revealed that the number of differentially expressed genes (DEGs) in the combined exposure group (885 DEGs) was lower than in the single exposure groups (BMDM: 1870 DEGs and PS: 9109 DEGs). Transcriptomic profiling indicated that individual stressors of BMDM and PS disrupted 113 and 123 pathways, respectively, predominantly associated with protein synthesis and energy metabolism. Conversely, combined exposure significantly enriched 86 pathways, including ribosome function and oxidative phosphorylation, thereby manifesting an antagonistic effect. This study provides new insights into the effects of BMDM and PS on Chlorella sp. and offers valuable information for the risk assessment of multiple pollutants.
Full article
(This article belongs to the Special Issue Effects of Acute Exposure to Toxicants on Oxidative Stress in Aquatic Organisms, 2nd Edition)
►▼
Show Figures

Graphical abstract

Journal Menu
► ▼ Journal Menu-
- Toxics Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Antioxidants, JoX, Metabolites, Molecules, Toxics, Veterinary Sciences, IJMS, Biomolecules
Recent Advances in Veterinary Pharmacology and Toxicology
Topic Editors: Chongshan Dai, Jichang LiDeadline: 1 December 2025
Topic in
Atmosphere, Buildings, Energies, Sustainability, Toxics
Indoor Air Quality and Built Environment
Topic Editors: Shen Yang, Grzegorz Majewski, Delia D'Agostino, Jianbang XiangDeadline: 31 December 2025
Topic in
Applied Sciences, Toxics, IJERPH, Biology, Cancers, Radiation
Disease Risks from Environmental Radiological Exposure
Topic Editors: Valentina Venuti, Francesco CaridiDeadline: 1 April 2026
Topic in
Clean Technol., Sustainability, Toxics, Water, Environments, Agriculture
Soil/Sediment Remediation and Wastewater Treatment
Topic Editors: Francesco Bianco, Silvio Matassa, Armando OlivaDeadline: 30 April 2026

Conferences
Special Issues
Special Issue in
Toxics
Toxicity and Safety Assessment of Exposure to Heavy Metals
Guest Editors: Cristina Carvalho, Michael AschnerDeadline: 14 November 2025
Special Issue in
Toxics
From Low-Tier to Individual Effects of Emerging Pollutants: Integrative Approaches to Ecotoxicological Assessments—2nd Edition
Guest Editors: Tiago Simoes, Hugo Monteiro, Nuno FerreiraDeadline: 15 November 2025
Special Issue in
Toxics
Toxicity Assessment and Safety Management of Nanomaterials
Guest Editor: Guotao PengDeadline: 20 November 2025
Special Issue in
Toxics
Plant Responses to Heavy Metal
Guest Editor: Yanqun ZuDeadline: 20 November 2025
Topical Collections
Topical Collection in
Toxics
Exposure and Effects of Environmental Pollution on Vulnerable Populations
Collection Editors: Matteo Vitali, Carmela Protano, Arianna Antonucci
Topical Collection in
Toxics
Artificial Intelligence and Data Mining for Toxicological Sciences
Collection Editors: Emilio Benfenati, Noel Aquilina










