Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism
Abstract
1. Introduction
2. Materials and Methods
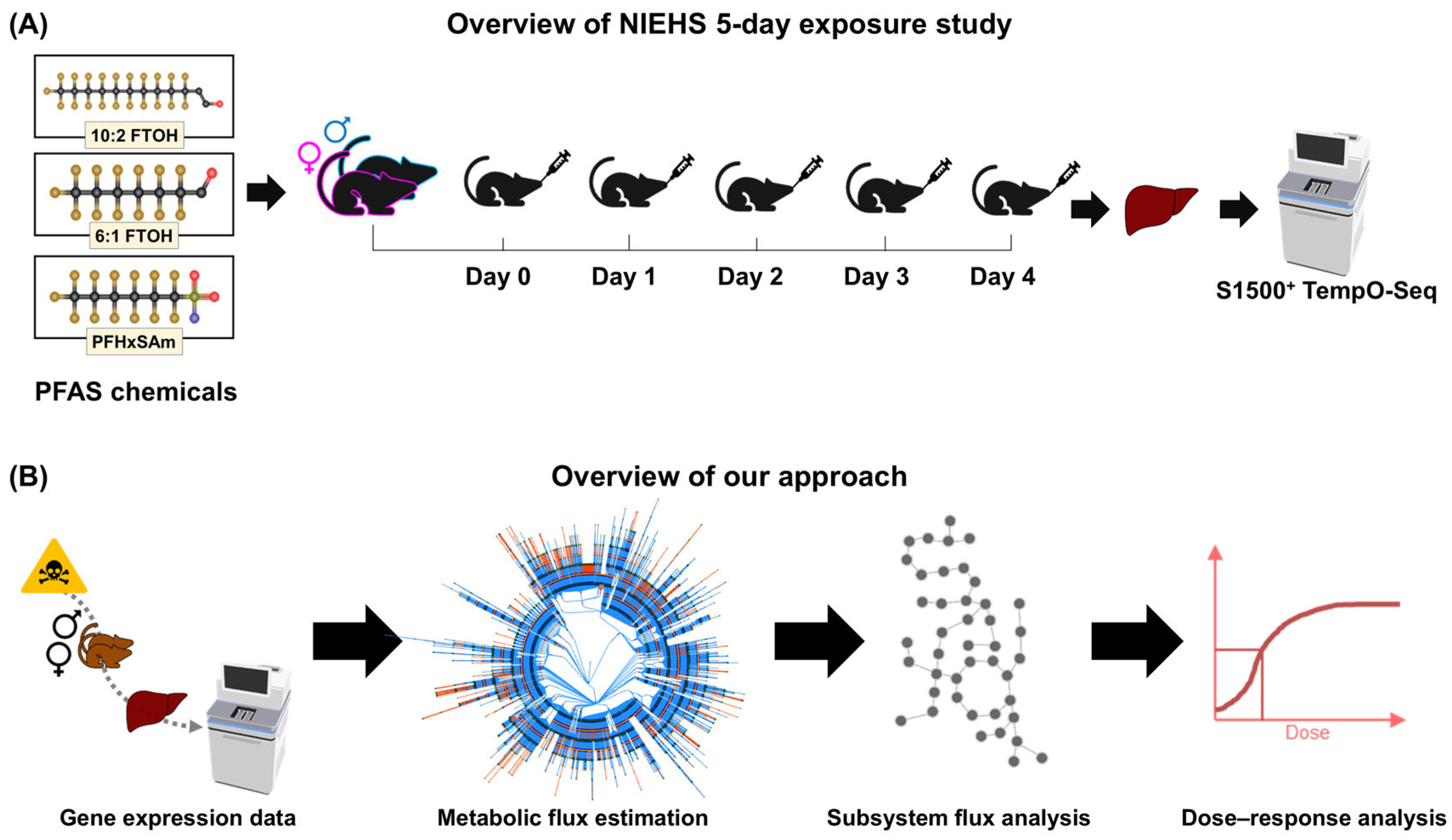
2.1. Animal Exposure Experiments and Transcriptomics
2.2. Computational Approach for Metabolic Risk Assessment of PFAS Chemicals
2.3. Rat Genome-Scale Metabolic Model
2.4. Fluxome Prediction Using the Pheflux Algorithm
2.5. Principal Component Analysis of Subsystem Fluxes
2.6. Computational Resources
2.7. Benchmark Dose Analysis
3. Results
3.1. Metabolic Flux Analysis to Quantify Liver Metabolic Activity Using Gene Expression Data
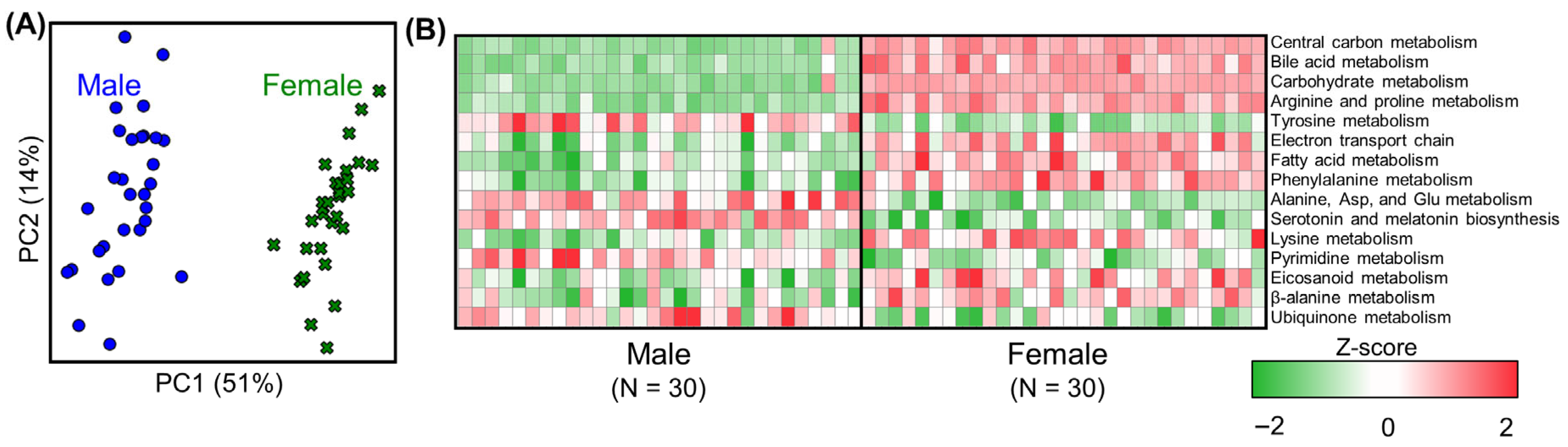
3.2. Metabolic Analysis of Liver Metabolism in Untreated Rats (Controls)
3.3. Effect of PFAS Exposure on Sexual Dimorphism in Male and Female Rat Livers
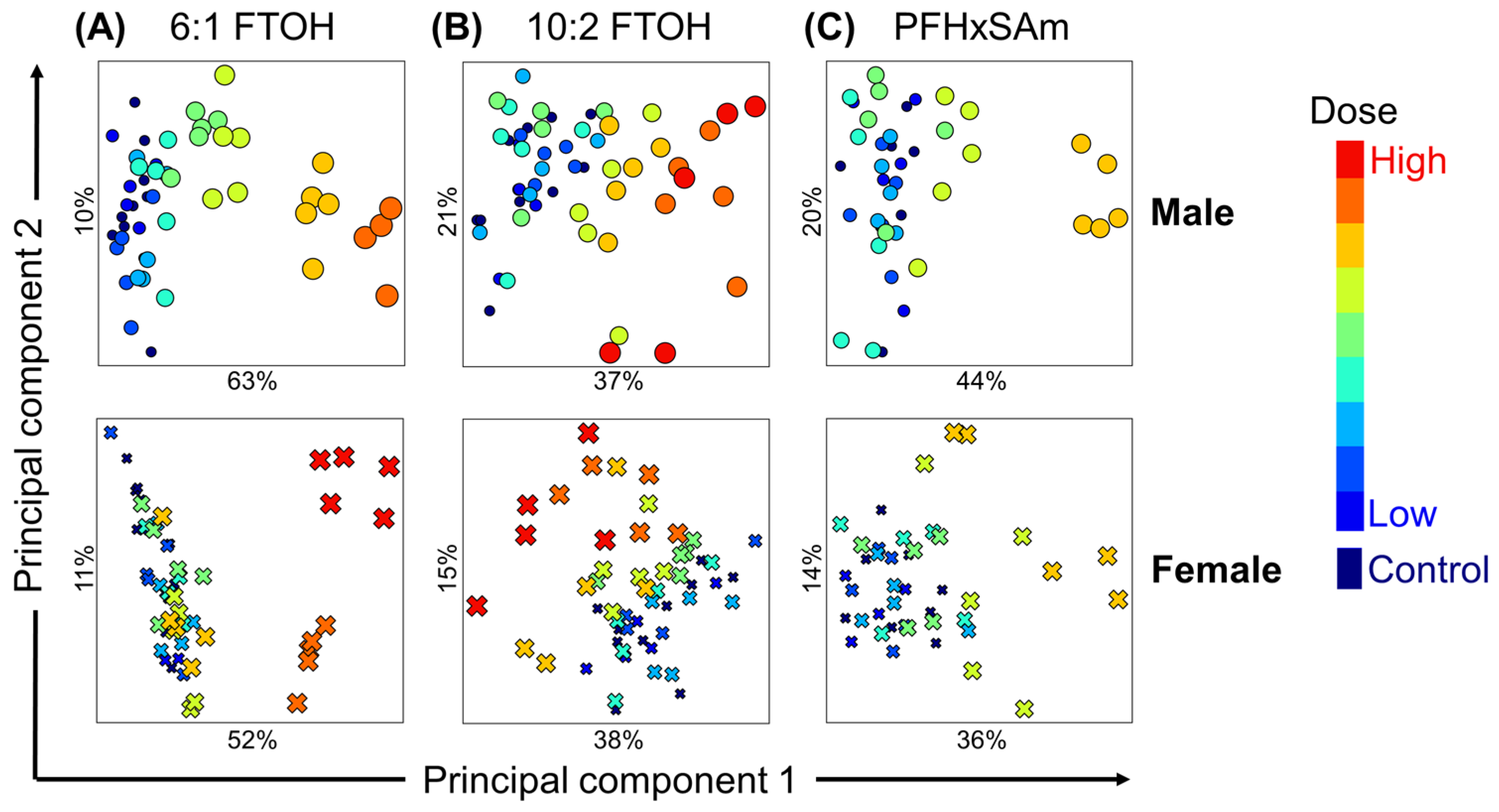
3.4. Analysis of PFAS Dose-Dependent Alterations in Rat Liver Metabolism
3.5. Metabolic Pathways Affected by PFAS Chemicals
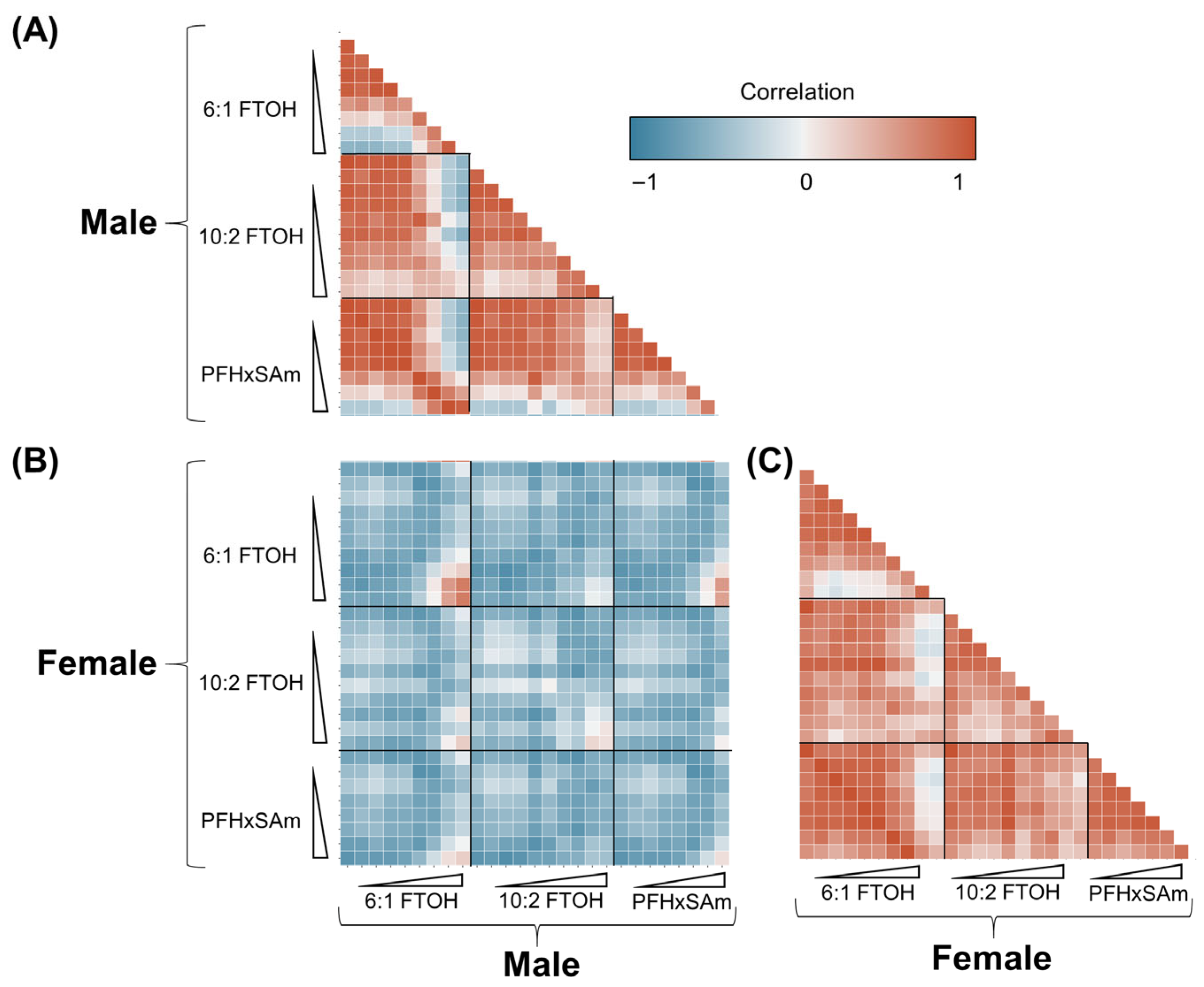
3.6. Correlation Between Male and Female Responses to PFAS Exposures
3.7. Benchmark Doses of PFAS Common Metabolic Alterations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PFASs | Per- and polyfluoroalkyl substances |
| GEM | Genome-scale metabolic model |
| BMD | Benchmark dose |
| PFCA | Perfluoroalkyl carboxylic acid |
| PFSA | Perfluoroalkyl sulfonate |
| PFOA | Perfluorooctanoic acid |
| PFOS | Perfluorooctanesulfonic acid |
| NAFLD | Non-alcoholic fatty liver disease |
| GPR | Gene–protein–reaction |
| EPA | Environmental Protection Agency |
| BMDS | Benchmark dose software |
| NIEHS | National Institute of Environmental Health Sciences |
| FTOH | Fluorotelomer alcohol |
| PFHxSAm | Perfluorohexanesulfonamide |
| PCA | Principal component analysis |
| BMR | Benchmark response |
| SD | Standard deviation |
| PC | Principal component |
| NASH | Non-alcoholic steatohepatitis |
| ROS | Reactive oxygen species |
References
- Sunderland, E.M.; Hu, X.C.; Dassuncao, C.; Tokranov, A.K.; Wagner, C.C.; Allen, J.G. A review of the pathways of human exposure to poly- and perfluoroalkyl substances (PFASs) and present understanding of health effects. J. Expo. Sci. Environ. Epidemiol. 2019, 29, 131–147. [Google Scholar] [CrossRef]
- Wang, Z.; DeWitt, J.C.; Higgins, C.P.; Cousins, I.T. A never-ending story of per- and polyfluoroalkyl substances (PFASs)? Environ. Sci. Technol. 2017, 51, 2508–2518. [Google Scholar] [CrossRef]
- Fenton, S.E.; Ducatman, A.; Boobis, A.; DeWitt, J.C.; Lau, C.; Ng, C.; Smith, J.S.; Roberts, S.M. Per- and polyfluoroalkyl substance toxicity and human health review: Current state of knowledge and strategies for informing future research. Environ. Toxicol. Chem. 2021, 40, 606–630. [Google Scholar] [CrossRef]
- Qi, W.; Clark, J.M.; Timme-Laragy, A.R.; Park, Y. Per- and polyfluoroalkyl substances and obesity, type 2 diabetes and non-alcoholic fatty liver disease: A review of epidemiologic findings. Toxicol. Environ. Chem. 2020, 102, 1–36. [Google Scholar] [CrossRef]
- Gluge, J.; Scheringer, M.; Cousins, I.T.; DeWitt, J.C.; Goldenman, G.; Herzke, D.; Lohmann, R.; Ng, C.A.; Trier, X.; Wang, Z. An overview of the uses of per- and polyfluoroalkyl substances (PFAS). Environ. Sci. Process Impacts 2020, 22, 2345–2373. [Google Scholar] [CrossRef] [PubMed]
- Buck, R.C.; Franklin, J.; Berger, U.; Conder, J.M.; Cousins, I.T.; de Voogt, P.; Jensen, A.A.; Kannan, K.; Mabury, S.A.; van Leeuwen, S.P. Perfluoroalkyl and polyfluoroalkyl substances in the environment: Terminology, classification, and origins. Integr. Environ. Assess. Manag. 2011, 7, 513–541. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Oliva, P.; Zhang, L.; Goodrich, J.A.; McConnell, R.; Conti, D.V.; Chatzi, L.; Aung, M. Associations between per-and polyfluoroalkyl substances (PFAS) and county-level cancer incidence between 2016 and 2021 and incident cancer burden attributable to PFAS in drinking water in the United States. J. Expo. Sci. Environ. Epidemiol. 2025, 35, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Huang, S.; Chen, X.; Huo, Q.; Xie, N.; Xia, J. Liver tissue metabolic profiling and pathways of non-alcoholic steatohepatitis in rats. Hepatol. Res. 2017, 47, 1484–1493. [Google Scholar] [CrossRef]
- Li, X.; Hou, M.; Zhang, F.; Ji, Z.; Cai, Y.; Shi, Y. Per- and polyfluoroalkyl substances and female health concern: Gender-based accumulation differences, adverse outcomes, and mechanisms. Environ. Sci. Technol. 2025, 59, 1469–1486. [Google Scholar] [CrossRef]
- Wackett, L.P. Confronting PFAS persistence: Enzymes catalyzing C-F bond cleavage. Trends Biochem. Sci. 2024, 50, 71–83. [Google Scholar] [CrossRef]
- Mišľanová, C.; Valachovičová, M. Health impacts of per- and polyfluoroalkyl substances (PFASs): A comprehensive review. Life 2025, 15, 573. [Google Scholar] [CrossRef]
- Wee, S.Y.; Aris, A.Z. Revisiting the “forever chemicals”, PFOA and PFOS exposure in drinking water. npj Clean. Water 2023, 6, 57. [Google Scholar] [CrossRef]
- Haug, M.; Dunder, L.; Lind, P.M.; Lind, L.; Salihovic, S. Associations of perfluoroalkyl substances (PFAS) with lipid and lipoprotein profiles. J. Expo. Sci. Environ. Epidemiol. 2023, 33, 757–765. [Google Scholar] [CrossRef]
- Sodani, K.; Ter Braak, B.; Hartvelt, S.; Boelens, M.; Jamalpoor, A.; Mukhi, S. Toxicological mode-of-action and developmental toxicity of different carbon chain length PFAS. Toxicol. Lett. 2025, 405, 59–66. [Google Scholar] [CrossRef]
- Wang, Z.; Cousins, I.T.; Scheringer, M.; Hungerbuehler, K. Hazard assessment of fluorinated alternatives to long-chain perfluoroalkyl acids (PFAAs) and their precursors: Status quo, ongoing challenges and possible solutions. Environ. Int. 2015, 75, 172–179. [Google Scholar] [CrossRef]
- Balgooyen, S.; Remucal, C.K. Impacts of environmental and engineered processes on the PFAS fingerprint of fluorotelomer-based AFFF. Environ. Sci. Technol. 2023, 57, 244–254. [Google Scholar] [CrossRef]
- Kolanczyk, R.C.; Saley, M.R.; Serrano, J.A.; Daley, S.M.; Tapper, M.A. PFAS biotransformation pathways: A species comparison study. Toxics 2023, 11, 74. [Google Scholar] [CrossRef]
- Bline, A.P.; DeWitt, J.C.; Kwiatkowski, C.F.; Pelch, K.E.; Reade, A.; Varshavsky, J.R. Public health risks of PFAS-related immunotoxicity are real. Curr. Environ. Health Rep. 2024, 11, 118–127. [Google Scholar] [CrossRef]
- Qiao, W.; Li, J.; Luo, L.; Peng, W.; Wang, X.; Jin, R.; Li, J. Triglycerides mediate the relationships of per- and poly-fluoroalkyl substance (PFAS) exposure with nonalcoholic fatty liver disease (NAFLD) risk in US participants. Ecotoxicol. Environ. Saf. 2024, 289, 117436. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, L.; Ducatman, A.; Deng, C.; von Stackelberg, K.E.; Danford, C.J.; Zhang, X. Association of per- and polyfluoroalkyl substance exposure with fatty liver disease risk in US adults. JHEP Rep. 2023, 5, 100694. [Google Scholar] [CrossRef]
- Sen, P.; Qadri, S.; Luukkonen, P.K.; Ragnarsdottir, O.; McGlinchey, A.; Jantti, S.; Juuti, A.; Arola, J.; Schlezinger, J.J.; Webster, T.F.; et al. Exposure to environmental contaminants is associated with altered hepatic lipid metabolism in non-alcoholic fatty liver disease. J. Hepatol. 2022, 76, 283–293. [Google Scholar] [CrossRef]
- Goodrich, J.A.; Walker, D.; Lin, X.; Wang, H.; Lim, T.; McConnell, R.; Conti, D.V.; Chatzi, L.; Setiawan, V.W. Exposure to perfluoroalkyl substances and risk of hepatocellular carcinoma in a multiethnic cohort. JHEP Rep. 2022, 4, 100550. [Google Scholar] [CrossRef]
- Bassler, J.; Ducatman, A.; Elliott, M.; Wen, S.; Wahlang, B.; Barnett, J.; Cave, M.C. Environmental perfluoroalkyl acid exposures are associated with liver disease characterized by apoptosis and altered serum adipocytokines. Environ. Pollut. 2019, 247, 1055–1063. [Google Scholar] [CrossRef]
- Maerten, A.; Callewaert, E.; Sanz-Serrano, J.; Devisscher, L.; Vinken, M. Effects of per- and polyfluoroalkyl substances on the liver: Human-relevant mechanisms of toxicity. Sci. Total Environ. 2024, 954, 176717. [Google Scholar] [CrossRef]
- Wan, H.T.; Zhao, Y.G.; Wei, X.; Hui, K.Y.; Giesy, J.P.; Wong, C.K. PFOS-induced hepatic steatosis, the mechanistic actions on beta-oxidation and lipid transport. Biochim. Biophys. Acta 2012, 1820, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Bagley, B.D.; Chang, S.C.; Ehresman, D.J.; Eveland, A.; Zitzow, J.D.; Parker, G.A.; Peters, J.M.; Wallace, K.B.; Butenhoff, J.L. Perfluorooctane sulfonate-induced hepatic steatosis in male Sprague Dawley rats is not attenuated by dietary choline supplementation. Toxicol. Sci. 2017, 160, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.; Rock, S.; Stratakis, N.; Eckel, S.P.; Walker, D.I.; Valvi, D.; Cserbik, D.; Jenkins, T.; Xanthakos, S.A.; Kohli, R.; et al. Exposure to per- and polyfluoroalkyl substances and markers of liver injury: A systematic review and meta-analysis. Environ. Health Perspect. 2022, 130, 46001. [Google Scholar] [CrossRef]
- Ducatman, A.; Fenton, S.E. Invited perspective: PFAS and liver disease: Bringing all the evidence together. Environ. Health Perspect. 2022, 130, 41303. [Google Scholar] [CrossRef]
- Roth, K.; Yang, Z.; Agarwal, M.; Liu, W.; Peng, Z.; Long, Z.; Birbeck, J.; Westrick, J.; Liu, W.; Petriello, M.C. Exposure to a mixture of legacy, alternative, and replacement per- and polyfluoroalkyl substances (PFAS) results in sex-dependent modulation of cholesterol metabolism and liver injury. Environ. Int. 2021, 157, 106843. [Google Scholar] [CrossRef]
- Kim, H.S.; Kwack, S.J.; Han, E.S.; Kang, T.S.; Kim, S.H.; Han, S.Y. Induction of apoptosis and CYP4A1 expression in Sprague-Dawley rats exposed to low doses of perfluorooctane sulfonate. J. Toxicol. Sci. 2011, 36, 201–210. [Google Scholar] [CrossRef]
- Schildroth, S.; Bond, J.C.; Wesselink, A.K.; Abrams, J.; Calafat, A.M.; Cook Botelho, J.; White, K.O.; Wegienka, G.; Hatch, E.E.; Wise, L.A. Associations between per- and polyfluoroalkyl substances (PFAS) and female sexual function in a preconception cohort. Environ. Res. 2024, 266, 120556. [Google Scholar] [CrossRef]
- Hari, A.; AbdulHameed, M.D.M.; Balik-Meisner, M.R.; Mav, D.; Phadke, D.P.; Scholl, E.H.; Shah, R.R.; Casey, W.; Auerbach, S.S.; Wallqvist, A.; et al. Exposure to PFAS chemicals induces sex-dependent alterations in key rate-limiting steps of lipid metabolism in liver steatosis. Front. Toxicol. 2024, 6, 1390196. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Singh, R.; Pasricha, C.; Kumari, P. Navigating liver health with metabolomics: A comprehensive review. Clin. Chim. Acta 2025, 566, 120038. [Google Scholar] [CrossRef]
- Chen, M. Environmental chemical exposomics and metabolomics in toxicology: The latest updates. Toxics 2024, 12, 647. [Google Scholar] [CrossRef]
- Hernandez-Mesa, M.; Le Bizec, B.; Dervilly, G. Metabolomics in chemical risk analysis—A review. Anal. Chim. Acta 2021, 1154, 338298. [Google Scholar] [CrossRef]
- Bouhifd, M.; Andersen, M.E.; Baghdikian, C.; Boekelheide, K.; Crofton, K.M.; Fornace, A.J., Jr.; Kleensang, A.; Li, H.; Livi, C.; Maertens, A.; et al. The human toxome project. ALTEX 2015, 32, 112–124. [Google Scholar] [CrossRef]
- Tsouka, S.; Masoodi, M. Metabolic pathway analysis: Advantages and pitfalls for the functional interpretation of metabolomics and lipidomics data. Biomolecules 2023, 13, 244. [Google Scholar] [CrossRef]
- Yarici, M.; Canturk, F.; Dursun, S.; Aydin, H.N.; Karabekmez, M.E. RSEA: A web server for pathway enrichment analysis of metabolic reaction sets. Biotechnol. Bioeng. 2025, 122, 2251–2258. [Google Scholar] [CrossRef]
- Pandey, V. MiNEApy: Enhancing enrichment network analysis in metabolic networks. Bioinformatics 2025, 41, btaf077. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Gu, C.; Kim, G.B.; Kim, W.J.; Kim, H.U.; Lee, S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019, 20, 121. [Google Scholar] [CrossRef]
- Carter, E.L.; Constantinidou, C.; Alam, M.T. Applications of genome-scale metabolic models to investigate microbial metabolic adaptations in response to genetic or environmental perturbations. Brief. Bioinform. 2023, 25, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Foguet, C.; Xu, Y.; Ritchie, S.C.; Lambert, S.A.; Persyn, E.; Nath, A.P.; Davenport, E.E.; Roberts, D.J.; Paul, D.S.; Di Angelantonio, E.; et al. Genetically personalised organ-specific metabolic models in health and disease. Nat. Commun. 2022, 13, 7356. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Agren, R.; Kampf, C.; Asplund, A.; Uhlen, M.; Nielsen, J. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 3083. [Google Scholar] [CrossRef]
- Moskon, M.; Rezen, T. Context-specific genome-scale metabolic modelling and its application to the analysis of COVID-19 metabolic signatures. Metabolites 2023, 13, 126. [Google Scholar] [CrossRef]
- Pannala, V.R.; Hari, A.; AbdulHameed, M.D.; Balik-Meisner, M.R.; Mav, D.; Phadke, D.P.; Scholl, E.H.; Shah, R.R.; Auerbach, S.S.; Wallqvist, A. Quantifying liver-toxic responses from dose-dependent chemical exposures using a rat genome-scale metabolic model. Toxicol. Sci. 2025, 204, 154–168. [Google Scholar] [CrossRef]
- Pannala, V.R.; Vinnakota, K.C.; Estes, S.K.; Trenary, I.; O’Brien, T.P.; Printz, R.L.; Papin, J.A.; Reifman, J.; Oyama, T.; Shiota, M.; et al. Genome-scale model-based identification of metabolite indicators for early detection of kidney toxicity. Toxicol. Sci. 2020, 173, 293–312. [Google Scholar] [CrossRef]
- Pannala, V.R.; Vinnakota, K.C.; Rawls, K.D.; Estes, S.K.; O’Brien, T.P.; Printz, R.L.; Papin, J.A.; Reifman, J.; Shiota, M.; Young, J.D.; et al. Mechanistic identification of biofluid metabolite changes as markers of acetaminophen-induced liver toxicity in rats. Toxicol. Appl. Pharmacol. 2019, 372, 19–32. [Google Scholar] [CrossRef]
- Rawls, K.D.; Blais, E.M.; Dougherty, B.V.; Vinnakota, K.C.; Pannala, V.R.; Wallqvist, A.; Kolling, G.L.; Papin, J.A. Genome-scale characterization of toxicity induced metabolic alterations in primary hepatocytes. Toxicol. Sci. 2019, 172, 279–291. [Google Scholar] [CrossRef]
- Vinnakota, K.C.; Pannala, V.R.; Wall, M.L.; Rahim, M.; Estes, S.K.; Trenary, I.; O’Brien, T.P.; Printz, R.L.; Reifman, J.; Shiota, M.; et al. Network modeling of liver metabolism to predict plasma metabolite changes during short-term fasting in the laboratory rat. Front. Physiol. 2019, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Guil, F.; Garcia, R.; Garcia, J.M. Adding metabolic tasks to Human GEM models to improve the study of gene targets and their associated toxicities. Sci. Rep. 2024, 14, 17265. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.J.; Holstege, C.P.; Papin, J.A. Metabolic modeling of sex-specific liver tissue suggests mechanism of differences in toxicological responses. PLoS Comput. Biol. 2023, 19, e1010927. [Google Scholar] [CrossRef]
- Wignall, J.A.; Shapiro, A.J.; Wright, F.A.; Woodruff, T.J.; Chiu, W.A.; Guyton, K.Z.; Rusyn, I. Standardizing benchmark dose calculations to improve science-based decisions in human health assessments. Environ. Health Perspect. 2014, 122, 499–505. [Google Scholar] [CrossRef]
- Davis, J.A.; Gift, J.S.; Zhao, Q.J. Introduction to benchmark dose methods and U.S. EPA’s benchmark dose software (BMDS) version 2.1.1. Toxicol. Appl. Pharmacol. 2011, 254, 181–191. [Google Scholar] [CrossRef]
- Phillips, J.R.; Svoboda, D.L.; Tandon, A.; Patel, S.; Sedykh, A.; Mav, D.; Kuo, B.; Yauk, C.L.; Yang, L.; Thomas, R.S.; et al. BMDExpress 2: Enhanced transcriptomic dose-response analysis workflow. Bioinformatics 2019, 35, 1780–1782. [Google Scholar] [CrossRef]
- Yang, L.; Allen, B.C.; Thomas, R.S. BMDExpress: A software tool for the benchmark dose analyses of genomic data. BMC Genom. 2007, 8, 387. [Google Scholar] [CrossRef]
- Sen, P.; Orešič, M. Integrating omics data in genome-scale metabolic modeling: A methodological perspective for precision medicine. Metabolites 2023, 13, 855. [Google Scholar] [CrossRef]
- González-Arrué, N.; Inostroza, I.; Conejeros, R.; Rivas-Astroza, M. Phenotype-specific estimation of metabolic fluxes using gene expression data. iScience 2023, 26, 106201. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, S.S.; Aillon, K.L.; Ballin, J.D.; Collins, B.J.; Cora, M.C.; Duncan, N.S.; Fostel, J.M.; Liu, Y.F.; Luh, J.; Machesky, N.J.; et al. NIEHS Report on the In Vivo Repeat Dose Biological Potency Study of 6:1 Fluorotelomer Alcohol (CASRN 375-82-6) in Sprague Dawley (Hsd:Sprague Dawley® SD®) Rats (Gavage Studies); NIEHS Report 07; NIEHS: Research Triangle Park, NC, USA, 2023. [Google Scholar] [CrossRef]
- Auerbach, S.S.; Ballin, J.D.; Blake, J.C.; Browning, D.B.; Collins, B.J.; Cora, M.C.; Fernando, R.A.; Fostel, J.M.; Liu, Y.F.; Luh, J.; et al. NIEHS Report on the In Vivo Repeat Dose Biological Potency Study of Perfluorohexanesulfonamide (CASRN 41997-13-1) in Sprague Dawley (Hsd:Sprague Dawley® SD®) Rats (Gavage Studies); NIEHS Report 10; NIEHS: Research Triangle Park, NC, USA, 2023. [Google Scholar] [CrossRef]
- Auerbach, S.S.; Ballin, J.D.; Blake, J.C.; Browning, D.B.; Collins, B.J.; Cora, M.C.; Fernando, R.A.; Fostel, J.M.; Liu, Y.F.; Luh, J.; et al. NIEHS Report on the In Vivo Repeat Dose Biological Potency Study of 1,1,2,2-Tetrahydroperfluoro-1-dodecanol (CASRN 865-86-1) in Sprague Dawley (Hsd:Sprague Dawley® SD®) Rats (Gavage Studies); NIEHS Report 08; NIEHS: Research Triangle Park, NC, USA, 2023. [Google Scholar] [CrossRef]
- Mav, D.; Shah, R.R.; Howard, B.E.; Auerbach, S.S.; Bushel, P.R.; Collins, J.B.; Gerhold, D.L.; Judson, R.S.; Karmaus, A.L.; Maull, E.A.; et al. A hybrid gene selection approach to create the S1500+ targeted gene sets for use in high-throughput transcriptomics. PLoS ONE 2018, 13, e0191105. [Google Scholar] [CrossRef]
- Mansouri, K.; Grulke, C.M.; Judson, R.S.; Williams, A.J. OPERA models for predicting physicochemical properties and environmental fate endpoints. J. Cheminform 2018, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Yeakley, J.M.; Shepard, P.J.; Goyena, D.E.; VanSteenhouse, H.C.; McComb, J.D.; Seligmann, B.E. A trichostatin A expression signature identified by TempO-Seq targeted whole transcriptome profiling. PLoS ONE 2017, 12, e0178302. [Google Scholar] [CrossRef]
- Sciome. GeniE. Available online: https://apps.sciome.com/genie/home (accessed on 31 July 2025).
- Hari, A.; Lobo, D. Fluxer: A web application to compute, analyze and visualize genome-scale metabolic flux networks. Nucleic Acids Res. 2020, 48, W427–W435. [Google Scholar] [CrossRef]
- Pannala, V.R.; Wall, M.L.; Estes, S.K.; Trenary, I.; O’Brien, T.P.; Printz, R.L.; Vinnakota, K.C.; Reifman, J.; Shiota, M.; Young, J.D.; et al. Metabolic network-based predictions of toxicant-induced metabolite changes in the laboratory rat. Sci. Rep. 2018, 8, 11678. [Google Scholar] [CrossRef]
- Blais, E.M.; Rawls, K.D.; Dougherty, B.V.; Li, Z.I.; Kolling, G.L.; Ye, P.; Wallqvist, A.; Papin, J.A. Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat. Commun. 2017, 8, 14250. [Google Scholar] [CrossRef]
- Wang, H.; Robinson, J.L.; Kocabas, P.; Gustafsson, J.; Anton, M.; Cholley, P.E.; Huang, S.; Gobom, J.; Svensson, T.; Uhlen, M.; et al. Genome-scale metabolic network reconstruction of model animals as a platform for translational research. Proc. Natl. Acad. Sci. USA 2021, 118, e2102344118. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ebrahim, A.; Lerman, J.A.; Palsson, B.O.; Hyduke, D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 2013, 7, 74. [Google Scholar] [CrossRef]
- Pham, L.L.; Watford, S.; Friedman, K.P.; Wignall, J.; Shapiro, A.J. Python BMDS: A Python interface library and web application for the canonical EPA dose-response modeling software. Reprod. Toxicol. 2019, 90, 102–108. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Trang, P.N.; Kumar, A. Understanding PFAS toxicity through cell culture metabolomics: Current applications and future perspectives. Environ. Int. 2024, 186, 108620. [Google Scholar] [CrossRef]
- Hasegawa, T.; Iino, C.; Endo, T.; Mikami, K.; Kimura, M.; Sawada, N.; Nakaji, S.; Fukuda, S. Changed amino acids in NAFLD and liver fibrosis: A large cross-sectional study without influence of insulin resistance. Nutrients 2020, 12, 1450. [Google Scholar] [CrossRef]
- van den Berg, E.H.; Flores-Guerrero, J.L.; Gruppen, E.G.; de Borst, M.H.; Wolak-Dinsmore, J.; Connelly, M.A.; Bakker, S.J.L.; Dullaart, R.P.F. Non-alcoholic fatty liver disease and risk of incident type 2 diabetes: Role of circulating branched-chain amino acids. Nutrients 2019, 11, 705. [Google Scholar] [CrossRef]
- Gaggini, M.; Carli, F.; Rosso, C.; Buzzigoli, E.; Marietti, M.; Della Latta, V.; Ciociaro, D.; Abate, M.L.; Gambino, R.; Cassader, M.; et al. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology 2018, 67, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Mardinoglu, A.; Bjornson, E.; Zhang, C.; Klevstig, M.; Soderlund, S.; Stahlman, M.; Adiels, M.; Hakkarainen, A.; Lundbom, N.; Kilicarslan, M.; et al. Personal model-assisted identification of NAD(+) and glutathione metabolism as intervention target in NAFLD. Mol. Syst. Biol. 2017, 13, 916. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, K.; Zikova-Kloas, A.; Marx-Stoelting, P.; Braeuning, A. Metabolism-disrupting chemicals affecting the liver: Screening, testing, and molecular pathway identification. Int. J. Mol. Sci. 2023, 24, 2686. [Google Scholar] [CrossRef]
- Kashobwe, L.; Sadrabadi, F.; Braeuning, A.; Leonards, P.E.G.; Buhrke, T.; Hamers, T. In vitro screening of understudied PFAS with a focus on lipid metabolism disruption. Arch. Toxicol. 2024, 98, 3381–3395. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Cai, D.; Chen, Q.; Zhu, Z.; Zhang, S.; Wang, Z.; Hu, Z.; Shen, H.; Meng, Z. Hunting metabolic biomarkers for exposure to per- and polyfluoroalkyl substances: A review. Metabolites 2024, 14, 392. [Google Scholar] [CrossRef]
- Guo, H.; Chen, J.; Zhang, H.; Yao, J.; Sheng, N.; Li, Q.; Guo, Y.; Wu, C.; Xie, W.; Dai, J. Exposure to GenX and its novel analogs disrupts hepatic bile acid metabolism in male mice. Environ. Sci. Technol. 2022, 56, 6133–6143. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Klaassen, C.D.; Cheng, X. Alteration of bile acid and cholesterol biosynthesis and transport by perfluorononanoic acid (PFNA) in mice. Toxicol. Sci. 2018, 162, 225–233. [Google Scholar] [CrossRef]
- Vairetti, M.; Di Pasqua, L.G.; Cagna, M.; Richelmi, P.; Ferrigno, A.; Berardo, C. Changes in glutathione content in liver diseases: An update. Antioxidants 2021, 10, 364. [Google Scholar] [CrossRef]
- Hossain, M.M.; Nawaz, M.Z.; Dar, M.A.; Geng, A.; Alghamdi, H.A.; Han, S.; Zhu, D. Per- and polyfluoroalkyl substances (PFAS) exposure in biota and remediation strategies: Toxicological and biochemical perspectives. J. Hazard. Mater. Adv. 2025, 17, 100579. [Google Scholar] [CrossRef]
- Narasimhan, K.; Vaitheeswari; Choi, E.; Chandran, N.S.; Eriksson, J.G.; Bendt, A.K.; Torta, F.; Mir, S.A. Integrated analysis of per- and polyfluoroalkyl substances and plasma lipidomics profiles in multi-ethnic Asian subjects for exposome research. Environ. Health 2024, 23, 105. [Google Scholar] [CrossRef]
- India-Aldana, S.; Yao, M.; Midya, V.; Colicino, E.; Chatzi, L.; Chu, J.; Gennings, C.; Jones, D.P.; Loos, R.J.F.; Setiawan, V.W.; et al. PFAS exposures and the human metabolome: A systematic review of epidemiological studies. Curr. Pollut. Rep. 2023, 9, 510–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, D.; Yan, S.; Cui, J.; Liang, Y.; Ren, S. Adverse effects of perfluorooctane sulfonate on the liver and relevant mechanisms. Toxics 2022, 10, 265. [Google Scholar] [CrossRef]
- Alijagic, A.; Sinisalu, L.; Duberg, D.; Kotlyar, O.; Scherbak, N.; Engwall, M.; Oresic, M.; Hyotylainen, T. Metabolic and phenotypic changes induced by PFAS exposure in two human hepatocyte cell models. Environ. Int. 2024, 190, 108820. [Google Scholar] [CrossRef]
- Ding, S.; Li, G.; Fu, T.; Zhang, T.; Lu, X.; Li, N.; Geng, Q. Ceramides and mitochondrial homeostasis. Cell. Signal. 2024, 117, 111099. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Ouro, A.; Ala-Ibanibo, L.; Presa, N.; Delgado, T.C.; Martinez-Chantar, M.L. Sphingolipids in non-alcoholic fatty liver disease and hepatocellular carcinoma: Ceramide turnover. Int. J. Mol. Sci. 2019, 21, 40. [Google Scholar] [CrossRef] [PubMed]
- McGlinchey, A.; Sinioja, T.; Lamichhane, S.; Sen, P.; Bodin, J.; Siljander, H.; Dickens, A.M.; Geng, D.; Carlsson, C.; Duberg, D.; et al. Prenatal exposure to perfluoroalkyl substances modulates neonatal serum phospholipids, increasing risk of type 1 diabetes. Environ. Int. 2020, 143, 105935. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Bizzarri, M. Inositols in insulin signaling and glucose metabolism. Int. J. Endocrinol. 2018, 2018, 1968450. [Google Scholar] [CrossRef]
- Auger, C.; Alhasawi, A.; Contavadoo, M.; Appanna, V.D. Dysfunctional mitochondrial bioenergetics and the pathogenesis of hepatic disorders. Front. Cell Dev. Biol. 2015, 3, 40. [Google Scholar] [CrossRef]
- Tilg, H.; Moschen, A.R. Evolution of inflammation in nonalcoholic fatty liver disease: The multiple parallel hits hypothesis. Hepatology 2010, 52, 1836–1846. [Google Scholar] [CrossRef]
- Mantena, S.K.; King, A.L.; Andringa, K.K.; Eccleston, H.B.; Bailey, S.M. Mitochondrial dysfunction and oxidative stress in the pathogenesis of alcohol- and obesity-induced fatty liver diseases. Free Radic. Biol. Med. 2008, 44, 1259–1272. [Google Scholar] [CrossRef] [PubMed]
- Allameh, A.; Niayesh-Mehr, R.; Aliarab, A.; Sebastiani, G.; Pantopoulos, K. Oxidative stress in liver pathophysiology and disease. Antioxidants 2023, 12, 1653. [Google Scholar] [CrossRef] [PubMed]
- Jaara, H.S.; Torres, S. Mitochondrial ROS, a trigger for mitochondrial dysfunction and inflammasome activation and a therapeutic target in liver diseases. Explor. Dig. Dis. 2024, 3, 474–503. [Google Scholar] [CrossRef]
- Gormaz, J.G.; Rodrigo, R.; Videla, L.A.; Beems, M. Biosynthesis and bioavailability of long-chain polyunsaturated fatty acids in non-alcoholic fatty liver disease. Prog. Lipid Res. 2010, 49, 407–419. [Google Scholar] [CrossRef]
- Alba, M.M.; Ebright, B.; Hua, B.; Slarve, I.; Zhou, Y.; Jia, Y.; Louie, S.G.; Stiles, B.L. Eicosanoids and other oxylipins in liver injury, inflammation and liver cancer development. Front. Physiol. 2023, 14, 1098467. [Google Scholar] [CrossRef]
- Montefusco, D.; Lambert, J.; Anderson, A.; Allegood, J.; Cowart, L.A. Analysis of the sphingolipidome in NAFLD. Methods Mol. Biol. 2022, 2455, 279–303. [Google Scholar] [CrossRef]
- Montefusco, D.J.; Allegood, J.C.; Spiegel, S.; Cowart, L.A. Non-alcoholic fatty liver disease: Insights from sphingolipidomics. Biochem. Biophys. Res. Commun. 2018, 504, 608–616. [Google Scholar] [CrossRef]
- Pralhada Rao, R.; Vaidyanathan, N.; Rengasamy, M.; Mammen Oommen, A.; Somaiya, N.; Jagannath, M.R. Sphingolipid metabolic pathway: An overview of major roles played in human diseases. J. Lipids 2013, 2013, 178910. [Google Scholar] [CrossRef]
- Pani, A.; Giossi, R.; Menichelli, D.; Fittipaldo, V.A.; Agnelli, F.; Inglese, E.; Romandini, A.; Roncato, R.; Pintaudi, B.; Del Sole, F.; et al. Inositol and non-alcoholic fatty liver disease: A systematic review on deficiencies and supplementation. Nutrients 2020, 12, 3379. [Google Scholar] [CrossRef]
- Delli Bovi, A.P.; Marciano, F.; Mandato, C.; Siano, M.A.; Savoia, M.; Vajro, P. Oxidative stress in non-alcoholic fatty liver disease. Front. Med. 2021, 8, 595371. [Google Scholar] [CrossRef]
- Jain, R.B.; Ducatman, A. Serum concentrations of selected perfluoroalkyl substances for US females compared to males as they age. Sci. Total Environ. 2022, 842, 156891. [Google Scholar] [CrossRef]
- Rickard, B.P.; Rizvi, I.; Fenton, S.E. Per- and poly-fluoroalkyl substances (PFAS) and female reproductive outcomes: PFAS elimination, endocrine-mediated effects, and disease. Toxicology 2022, 465, 153031. [Google Scholar] [CrossRef] [PubMed]
- Della Torre, S.; Mitro, N.; Meda, C.; Lolli, F.; Pedretti, S.; Barcella, M.; Ottobrini, L.; Metzger, D.; Caruso, D.; Maggi, A. Short-term fasting reveals amino acid metabolism as a major sex-discriminating factor in the liver. Cell Metab. 2018, 28, 256–267.e5. [Google Scholar] [CrossRef]
- Justo, R.; Boada, J.; Frontera, M.; Oliver, J.; Bermudez, J.; Gianotti, M. Gender dimorphism in rat liver mitochondrial oxidative metabolism and biogenesis. Am. J. Physiol. Cell Physiol. 2005, 289, C372–C378. [Google Scholar] [CrossRef]
- Phelps, T.; Snyder, E.; Rodriguez, E.; Child, H.; Harvey, P. The influence of biological sex and sex hormones on bile acid synthesis and cholesterol homeostasis. Biol. Sex. Differ. 2019, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Zhou, Z.; Hu, Z.; Wei, C.; Li, J.; Wang, L.; Liu, G.; Zhang, J.; Wang, Y.; Wang, T.; et al. Effect of enterohepatic circulation on the accumulation of per- and polyfluoroalkyl substances: Evidence from experimental and computational studies. Environ. Sci. Technol. 2022, 56, 3214–3224. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, W.M.; Auerbach, S.S.; Parham, F.; Stout, M.D.; Waidyanatha, S.; Mutlu, E.; Collins, B.; Paules, R.S.; Merrick, B.A.; Ferguson, S.; et al. Evaluation of 5-day in vivo rat liver and kidney with high-throughput transcriptomics for estimating benchmark doses of apical outcomes. Toxicol. Sci. 2020, 176, 343–354. [Google Scholar] [CrossRef]
- Brunk, E.; Sahoo, S.; Zielinski, D.C.; Altunkaya, A.; Drager, A.; Mih, N.; Gatto, F.; Nilsson, A.; Preciat Gonzalez, G.A.; Aurich, M.K.; et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. 2018, 36, 272–281. [Google Scholar] [CrossRef]
- Beale, D.J.; Sinclair, G.M.; Shah, R.; Paten, A.M.; Kumar, A.; Long, S.M.; Vardy, S.; Jones, O.A.H. A review of omics-based PFAS exposure studies reveals common biochemical response pathways. Sci. Total Environ. 2022, 845, 157255. [Google Scholar] [CrossRef]
- Barouki, R.; Samson, M.; Blanc, E.B.; Colombo, M.; Zucman-Rossi, J.; Lazaridis, K.N.; Miller, G.W.; Coumoul, X. The exposome and liver disease—How environmental factors affect liver health. J. Hepatol. 2023, 79, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Manautou, J.E. Molecular mechanisms underlying chemical liver injury. Expert Rev. Mol. Med. 2012, 14, e4. [Google Scholar] [CrossRef] [PubMed]



| PFAS Chemical | CASRN | PubChem CID | OPERA LD50 Prediction (Uncertainty Range), mg/kg/Day | U.S. EPA Estimated POD (Uncertainty Range), mg/kg/Day | Selected Dose Levels, mg/kg |
|---|---|---|---|---|---|
| 6:1 FTOH | 375-82-6 | 550386 | 460 (230–918) | 85 (0.6–637) | 0, 0.15, 0.50, 1.40, 4, 12, 37, 111, 333, 1000 |
| 10:2 FTOH | 865-86-1 | 70083 | 636 (319–1270) | 18 (0.3–197) | 0, 0.07, 0.20, 0.70, 2, 6, 18, 55, 160, 475 |
| PFHxSAm | 41997-13-1 | 11603678 | 263 (131–525) | 35 (0.9–916) | 0, 0.15, 0.50, 1.40, 4, 12, 37, 111, 333, 1000 |
| Metabolic Subsystem | Male | Female | ||||
|---|---|---|---|---|---|---|
| 6:1 FTOH | 10:2 FTOH | PFHxSAm | 6:1 FTOH | 10:2 FTOH | PFHxSAm | |
| β-alanine metabolism * | 6.6 | 55.8 | 260.4 | 49.0 | ||
| Cysteine and methionine metabolism | 37.5 | 33.0 | 23.4 | 59.1 | ||
| Eicosanoid metabolism * | 7.7 | 66.2 | 54.6 | 312.9 | ||
| Electron transport chain * | 3.9 | 304.6 | ||||
| Fatty acid biosynthesis * | 4.8 | 12.9 | 251.7 | |||
| Fatty acid metabolism * | 4.0 | 298.8 | 317.5 | 63.0 | ||
| Fatty acid oxidation * | 2.3 | 21.9 | 20.6 | 107.8 | 191.2 | 31.0 |
| Glutathione metabolism | 39.9 | 10.1 | 20.0 | 10.5 | ||
| Inositol phosphate metabolism * | 4.3 | |||||
| Nucleotide metabolism | 4.0 | 11.8 | 76.8 | 22.3 | ||
| Omega-3 fatty acid metabolism | 22.5 | 75.5 | ||||
| Omega-6 fatty acid metabolism | 1.6 | 36.2 | 18.9 | 31.0 | ||
| Porphyrin metabolism | 9.6 | |||||
| Protein metabolism | 5.0 | |||||
| Purine metabolism | 2.0 | 9.8 | 8.5 | 36.2 | 10.0 | 7.4 |
| Serotonin and melatonin biosynthesis | 17.7 | 22.5 | 618.9 | 17.6 | ||
| Sphingolipid metabolism * | 2.9 | 13.1 | 35.1 | 12.2 | ||
| Tyrosine metabolism | 4.3 | 70.8 | 145.4 | 12.3 | ||
| Ubiquinone synthesis | 3.8 | 8.4 | 20.4 | 23.8 | 18.4 | |
| Valine, leucine, and isoleucine metabolism | 1.4 | 159.5 | 20.1 | |||
| Xenobiotic metabolism | 16.4 | 40.1 | 23.9 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hari, A.; Balik-Meisner, M.R.; Mav, D.; Phadke, D.P.; Scholl, E.H.; Shah, R.R.; Casey, W.; Auerbach, S.S.; Wallqvist, A.; Pannala, V.R. Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism. Toxics 2025, 13, 684. https://doi.org/10.3390/toxics13080684
Hari A, Balik-Meisner MR, Mav D, Phadke DP, Scholl EH, Shah RR, Casey W, Auerbach SS, Wallqvist A, Pannala VR. Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism. Toxics. 2025; 13(8):684. https://doi.org/10.3390/toxics13080684
Chicago/Turabian StyleHari, Archana, Michele R. Balik-Meisner, Deepak Mav, Dhiral P. Phadke, Elizabeth H. Scholl, Ruchir R. Shah, Warren Casey, Scott S. Auerbach, Anders Wallqvist, and Venkat R. Pannala. 2025. "Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism" Toxics 13, no. 8: 684. https://doi.org/10.3390/toxics13080684
APA StyleHari, A., Balik-Meisner, M. R., Mav, D., Phadke, D. P., Scholl, E. H., Shah, R. R., Casey, W., Auerbach, S. S., Wallqvist, A., & Pannala, V. R. (2025). Genome-Scale Metabolic Modeling Predicts Per- and Polyfluoroalkyl Substance-Mediated Early Perturbations in Liver Metabolism. Toxics, 13(8), 684. https://doi.org/10.3390/toxics13080684









