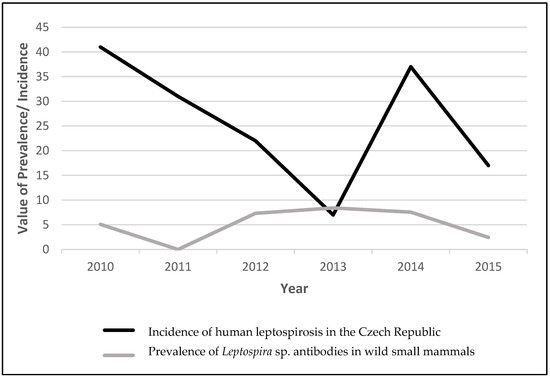
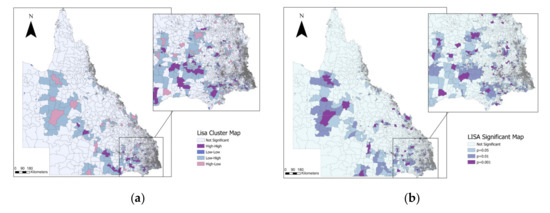
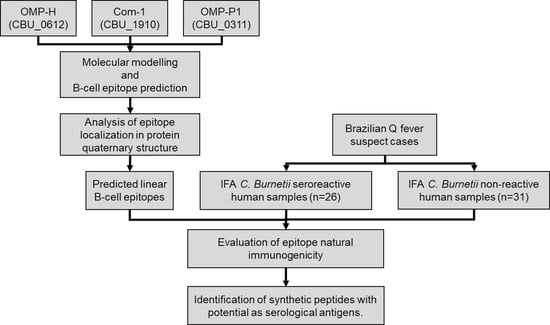
Updates on Rickettsia and Coxiella
A topical collection in Pathogens (ISSN 2076-0817). This collection belongs to the section "Parasitic Pathogens".
Submission Status: Closed | Viewed by 116272Editors
Interests: molecular diagnostics; microbial molecular biology; Q fever; Rickettsia infections; Rickettsiaceae; Coxiella burnetii; microbiology; ticks; zoonotic diseases; DNA
2. Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Thailand
Interests: rickettsial organisms; tropical diseases; global health; arthropod-borne pathogens; zoonotic diseases; diagnostics development
Topical Collection Information
Dear Colleagues,
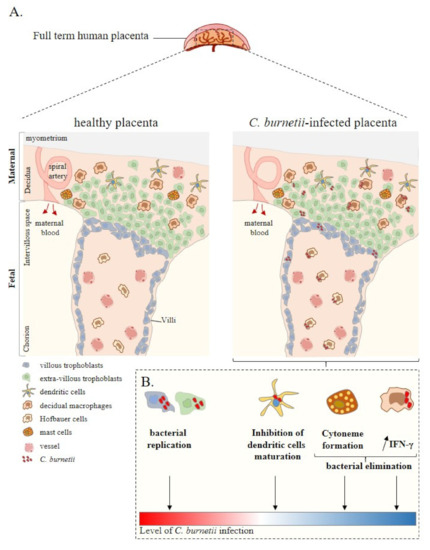
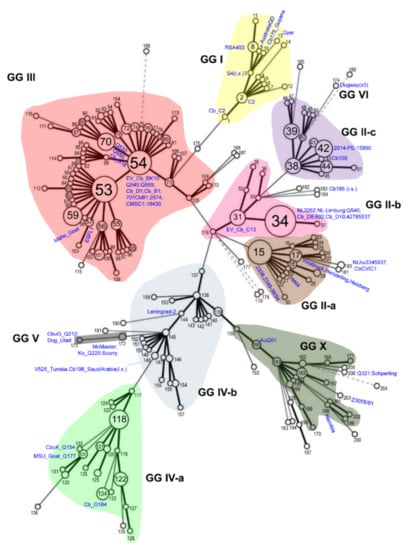
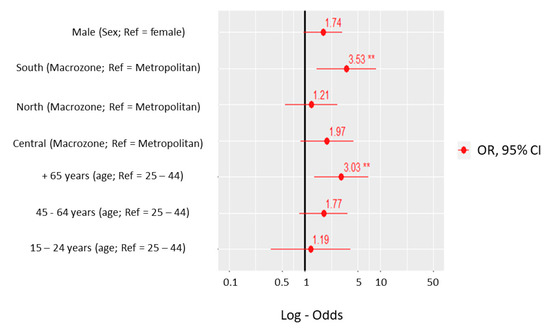
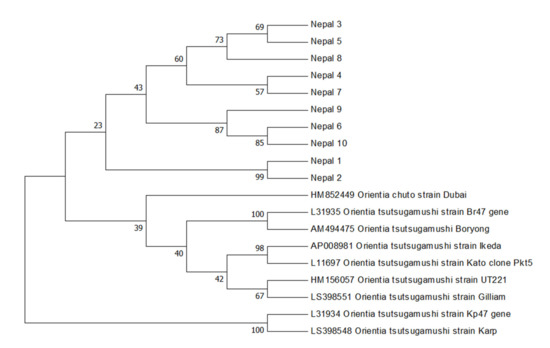
Rickettsiae and Coxiella are distinct bacterial groups, with the former an alphaproteobacterium and the later a gammaproteopbacterium, each with unique growth dynamics. Rickettsiae replicate in the cytosol or nucleus of eukaryotic cells while Coxiella grow in a pathogen-modified phagolysosome. These differences extend to their routes of transmission with rickettsiae mostly transmitted by arthropod vectors and Coxiella by air-borne transmission. However there is significant overlap in numerous aspects of their biology. This includes overlapping environmental niches, host and vector animals, and infection profiles in humans.
This Topic Collection, published by the journal Pathogens, will provide our small but productive community with an update on Coxiella and Rickettsiae. We are seeking papers from a wide scope, including epidemiological, species description and characterization, diagnostic assays, and vaccine development. We invite our colleagues from all disciplines, including clinical and basic research, to contribute their manuscripts for publication review in this Topic Collection of the journal dedicated to our favorite microbes.
Dr. John Stenos
Dr. Mohammad Yazid Abdad
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Pathogens is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2200 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Rickettsiae
- Coxiella