Identification of Immunogenic Linear B-Cell Epitopes in C. burnetii Outer Membrane Proteins Using Immunoinformatics Approaches Reveals Potential Targets of Persistent Infections
Abstract
:1. Introduction
2. Results
2.1. Prediction of Linear B-Cell Epitopes in C. burnetii OMPs
2.2. Prediction of Epitopes’ Antigenicity and Specificity
2.3. Epitope Exposition in Protein Quaternary Structure
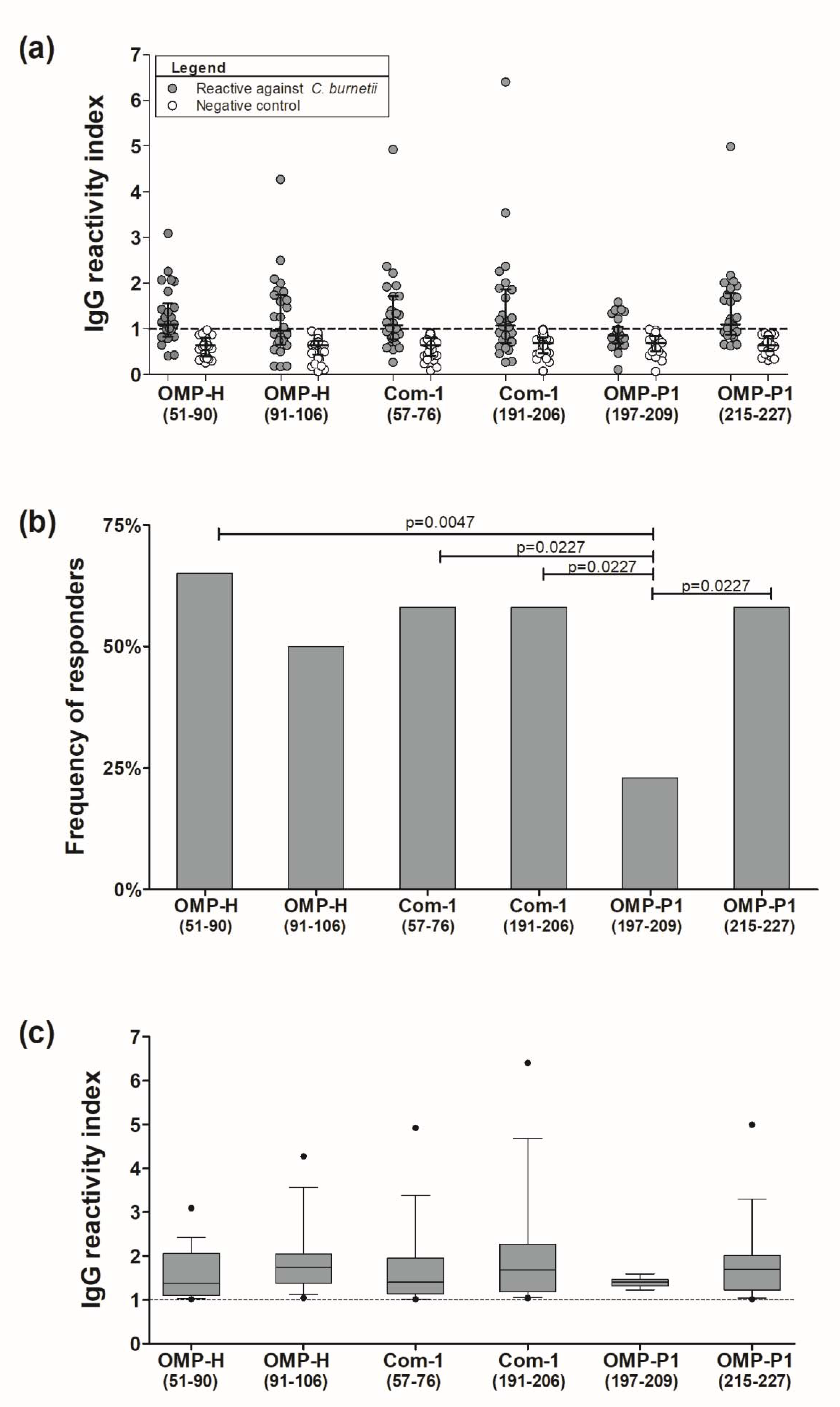
2.4. Studied Population Description
2.5. Preliminary Assessment of the Potential of Epitopes as Serological Antigens
2.6. Associations between Humoral Response to Synthetic Peptides and Clinical Features
3. Discussion
4. Materials and Methods
4.1. Sequence Data
4.2. B-Cell Epitope Prediction
4.3. Evaluation of B-Cell Epitopes’ Degree of Conservation
4.4. Three-Dimensional (3D) Structure Modeling
4.5. Evaluation of Epitopes’ Exposure in the Protein Quaternary Structures
4.6. Peptide Synthesis
4.7. Studied Population
4.8. Experimental Confirmation of the Antigenicity of the Predicted Epitopes
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Eldin, C.; Melenotte, C.; Mediannikov, O.; Ghigo, E.; Million, M.; Edouard, S.; Mege, J.L.; Maurin, M.; Raoult, D. From Q fever to Coxiella burnetii infection: A paradigm change. Clin. Microbiol. Rev. 2017, 30, 115–190. [Google Scholar] [CrossRef] [Green Version]
- ILRI. Mapping of Poverty and Likely Zoonoses Hotspots; Department for International Development, UK, 2012. Available online: https://cgspace.cgiar.org/bitstream/handle/10568/21161/ZooMap_July2012_final.pdf (accessed on 19 August 2021).
- Sahu, R.; Rawool, D.B.; Dhaka, P.; Yadav, J.P.; Mishra, S.P.; Kumar, M.; Vergis, J.; Malik, S.S.; Barbuddhe, S.B. Current perspectives on the occurrence of Q fever: Highlighting the need for systematic surveillance for a neglected zoonotic disease in Indian subcontinent. Environ. Microbiol. Rep. 2021, 13, 138–158. [Google Scholar] [CrossRef] [PubMed]
- Maurin, M.; Raoult, D. Q fever. Clin. Microbiol. Rev. 1999, 12, 518–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asamoah, J.K.K.; Jin, Z.; Sun, G.Q.; Li, M.Y. A deterministic model for Q fever transmission dynamics within dairy cattle herds: Using sensitivity analysis and optimal controls. Comput. Math. Methods Med. 2020, 2020, 6820608. [Google Scholar] [CrossRef] [PubMed]
- Rotz, L.D.; Khan, A.S.; Lillibridge, S.R.; Ostroff, S.M.; Hughes, J.M. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 2002, 8, 225–230. [Google Scholar] [CrossRef]
- Woldehiwet, Z. Q fever (coxiellosis): Epidemiology and pathogenesis. Res. Vet. Sci. 2004, 77, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.E.; Hubbard, K.R.; Johnson, A.J.; Messick, J.B.; Weng, H.Y.; Pogranichniy, R.M. A cross sectional study evaluating the prevalence of Coxiella burnetii, potential risk factors for infection, and agreement between diagnostic methods in goats in Indiana. Prev. Vet. Med. 2016, 126, 131–137. [Google Scholar] [CrossRef]
- Patil, S.M.; Regunath, H. Q Fever; StatPearls Publishing: Treasure Island, FL, USA, 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK556095/ (accessed on 19 August 2021).
- Fishbein, D.B.; Raoult, D. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am. J. Trop. Med. Hyg. 1992, 47, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Esmaeili, S.; Mobarez, A.M.; Khalili, M.; Mostafavi, E. High prevalence and risk factors of Coxiella burnetii in milk of dairy animals with a history of abortion in Iran. Comp. Immunol. Microbiol. Infect. Dis. 2019, 63, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Raoult, D. Emergence of Q fever. Iran. J. Public Health 2011, 40, 1–18. [Google Scholar]
- Van den Brom, R.; van Engelen, E.; Roest, H.I.; van der Hoek, W.; Vellema, P. Coxiella burnetii infections in sheep or goats: An opinionated review. Vet. Microbiol. 2015, 181, 119–129. [Google Scholar] [CrossRef] [PubMed]
- van Schaik, E.J.; Chen, C.; Mertens, K.; Weber, M.M.; Samuel, J.E. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 2013, 11, 561–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Million, M.; Thuny, F.; Richet, H.; Raoult, D. Long-term outcome of Q fever endocarditis: A 26-year personal survey. Lancet Infect. Dis. 2010, 10, 527–535. [Google Scholar] [CrossRef]
- Delsing, C.E.; Kullberg, B.J.; Bleeker-Rovers, C.P. Q fever in the Netherlands from 2007 to 2010. Neth. J. Med. 2010, 68, 382–387. [Google Scholar]
- Porter, S.R.; Czaplicki, G.; Mainil, J.; Guatteo, R.; Saegerman, C. Q fever: Current state of knowledge and perspectives of research of a neglected zoonosis. Int. J. Microbiol. 2011, 2011, 248418. [Google Scholar] [CrossRef] [PubMed]
- Marrie, T.J.; Raoult, D. Q fever—A review and issues for the next century. Int. J. Antimicrob. Agents 1997, 8, 145–161. [Google Scholar] [CrossRef]
- Anderson, A.; Bijlmer, H.; Fournier, P.E.; Graves, S.; Hartzell, J.; Kersh, G.J.; Limonard, G.; Marrie, T.J.; Massung, R.F.; McQuiston, J.H.; et al. Diagnosis and management of Q fever—United States, 2013: Recommendations from CDC and the Q fever working group. Morb. Mortal. Wkly. Rep. Recomm. Rep. 2013, 62, 1–30. [Google Scholar]
- Mares-Guia, M.A.; Rozental, T.; Guterres, A.; Gomes, R.; Almeida, D.N.; Moreira, N.S.; Barreira, J.D.; Favacho, A.R.; Santana, A.L.; Lemos, E.R. Molecular identification of the agent of Q fever—Coxiella burnetii—in domestic animals in State of Rio de Janeiro, Brazil. Rev. Soc. Bras. Med. Trop. 2014, 47, 231–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mioni, M.S.R.; Costa, F.B.; Ribeiro, B.L.D.; Teixeira, W.S.R.; Pelicia, V.C.; Labruna, M.B.; Rousset, E.; Sidi-Boumedine, K.; Thiery, R.; Megid, J. Coxiella burnetii in slaughterhouses in Brazil: A public health concern. PLoS ONE 2020, 15, e0241246. [Google Scholar] [CrossRef]
- Zanatto, D.C.S.; Gatto, I.R.H.; Labruna, M.B.; Jusi, M.M.G.; Samara, S.I.; Machado, R.Z.; Andre, M.R. Coxiella burnetii associated with BVDV (Bovine Viral Diarrhea Virus), BoHV (Bovine Herpesvirus), Leptospira spp., Neospora caninum, Toxoplasma gondii and Trypanosoma vivax in reproductive disorders in cattle. Rev. Bras. Parasitol. Vet. 2019, 28, 245–257. [Google Scholar] [CrossRef]
- Souza, E.A.R.; Castro, E.M.S.; Oliveira, G.M.B.; Azevedo, S.S.; Peixoto, R.M.; Labruna, M.B.; Horta, M.C. Serological diagnosis and risk factors for Coxiella burnetii in goats and sheep in a semi-arid region of Northeastern Brazil. Rev. Bras. Parasitol. Vet. 2018, 27, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Mares-Guia, M.A.; Rozental, T.; Guterres, A.; Ferreira Mdos, S.; Botticini Rde, G.; Terra, A.K.; Marraschi, S.; Bochner, R.; Lemos, E.R. Molecular identification of Q fever in patients with a suspected diagnosis of dengue in Brazil in 2013–2014. Am. J. Trop. Med. Hyg. 2016, 94, 1090–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro-Netto, A.; Nikitin, T.; Valentini, H.; Ribeiro, I.F. Study of Q fever in S Ao Paulo. I. Incidence in dairy cattle in the Para’ibo Valley. Rev. Inst. Med. Trop. Sao Paulo 1964, 6, 137–141. [Google Scholar]
- Mioni, M.S.R.; Sidi-Boumedine, K.; Dalanezi, F.M.; Joaquim, S.F.; Denadai, R.; Teixeira, W.S.R.; Labruna, M.B.; Megid, J. New genotypes of Coxiella burnetii circulating in Brazil and Argentina. Pathogens 2019, 9, 30. [Google Scholar] [CrossRef] [Green Version]
- Rozental, T.; Silva, A.; Oliveira, R.C.; Favacho, A.R.M.; Oliveira, M.L.A.; Bastos, F.I.; Lemos, E.R.S. Seroprevalence of bartonella spp., Coxiella burnetii, and Hantavirus among people who inject drugs in Rio de Janeiro, Brazil: A retrospective assessment of a biobank. Rev. Inst. Med. Trop. Sao Paulo 2018, 60, e31. [Google Scholar] [CrossRef]
- Rozental, T.; Faria, L.S.; Forneas, D.; Guterres, A.; Ribeiro, J.B.; Araujo, F.R.; Lemos, E.R.S.; Silva, M.R. First molecular detection of Coxiella burnetii in Brazilian artisanal cheese: A neglected food safety hazard in ready-to-eat raw-milk product. Braz. J. Infect. Dis. 2020, 24, 208–212. [Google Scholar] [CrossRef]
- de Souza Ribeiro Mioni, M.; Ribeiro, B.L.D.; Peres, M.G.; Teixeira, W.S.R.; Pelicia, V.C.; Motta, R.G.; Labruna, M.B.; Ribeiro, M.G.; Sidi-Boumedine, K.; Megid, J. Real-time quantitative PCR-based detection of Coxiella burnetii in unpasteurized cow’s milk sold for human consumption. Zoonoses Public Health 2019, 66, 695–700. [Google Scholar] [CrossRef]
- Vaidya, V.M.; Malik, S.V.; Bhilegaonkar, K.N.; Rathore, R.S.; Kaur, S.; Barbuddhe, S.B. Prevalence of Q fever in domestic animals with reproductive disorders. Comp. Immunol. Microbiol. Infect. Dis. 2010, 33, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Vincent, G.A.; Graves, S.R.; Robson, J.M.; Nguyen, C.; Hussain-Yusuf, H.; Islam, A.; Fenwick, S.G.; Stenos, J. Isolation of Coxiella burnetii from serum of patients with acute Q fever. J. Microbiol. Methods 2015, 119, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Madariaga, M.G.; Rezai, K.; Trenholme, G.M.; Weinstein, R.A. Q fever: A biological weapon in your backyard. Lancet Infect. Dis. 2003, 3, 709–721. [Google Scholar] [CrossRef]
- Fournier, P.E.; Marrie, T.J.; Raoult, D. Diagnosis of Q fever. J. Clin. Microbiol. 1998, 36, 1823–1834. [Google Scholar] [CrossRef] [Green Version]
- Stephen, S.; Ambroise, S.; Pradeep, J.; Gunasekaran, D.; Sangeetha, B.; Sarangapani, K. Unreliability of three commercial Coxiella burnetii phase II IgM ELISA kits for the seroscreening of acute Q fever in human cases. Indian J. Med Res. 2017, 146, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Kowalczewska, M.; Nappez, C.; Vincentelli, R.; La Scola, B.; Raoult, D. Protein candidates for Q fever serodiagnosis. FEMS Immunol. Med. Microbiol. 2012, 64, 140–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beare, P.A.; Chen, C.; Bouman, T.; Pablo, J.; Unal, B.; Cockrell, D.C.; Brown, W.C.; Barbian, K.D.; Porcella, S.F.; Samuel, J.E.; et al. Candidate antigens for Q fever serodiagnosis revealed by immunoscreening of a Coxiella burnetii protein microarray. Clin. Vaccine Immunol. 2008, 15, 1771–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekeyova, Z.; Kowalczewska, M.; Decloquement, P.; Pelletier, N.; Spitalska, E.; Raoult, D. Identification of protein candidates for the serodiagnosis of Q fever endocarditis by an immunoproteomic approach. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 287–295. [Google Scholar] [CrossRef]
- Xiong, X.; Wang, X.; Wen, B.; Graves, S.; Stenos, J. Potential serodiagnostic markers for Q fever identified in Coxiella burnetii by immunoproteomic and protein microarray approaches. BMC Microbiol. 2012, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigil, A.; Ortega, R.; Nakajima-Sasaki, R.; Pablo, J.; Molina, D.M.; Chao, C.C.; Chen, H.W.; Ching, W.M.; Felgner, P.L. Genome-wide profiling of humoral immune response to Coxiella burnetii infection by protein microarray. Proteomics 2010, 10, 2259–2269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, J.; Xiong, X.; Qi, Y.; Gong, W.; Duan, C.; Yang, X.; Wen, B. Serological characterization of surface-exposed proteins of Coxiella burnetii. Microbiology 2014, 160, 2718–2731. [Google Scholar] [CrossRef]
- Skultety, L. Workshop on Q fever. Acta Virol. 2017, 61, 347–348. [Google Scholar] [CrossRef] [Green Version]
- Flores-Ramirez, G.; Danchenko, M.; Quevedo-Diaz, M.; Skultety, L. Reliable tool for detection of novel Coxiella burnetii antigens, using immobilized human polyclonal antibodies. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2017, 1047, 84–91. [Google Scholar] [CrossRef]
- Vranakis, I.; Mathioudaki, E.; Kokkini, S.; Psaroulaki, A. Com1 as a promising protein for the differential diagnosis of the two forms of Q fever. Pathogens 2019, 8, 242. [Google Scholar] [CrossRef] [Green Version]
- Sandoz, K.M.; Popham, D.L.; Beare, P.A.; Sturdevant, D.E.; Hansen, B.; Nair, V.; Heinzen, R.A. Transcriptional profiling of Coxiella burnetii reveals extensive cell wall remodeling in the small cell variant developmental form. PLoS ONE 2016, 11, e0149957. [Google Scholar] [CrossRef] [Green Version]
- Varghees, S.; Kiss, K.; Frans, G.; Braha, O.; Samuel, J.E. Cloning and porin activity of the major outer membrane protein P1 from Coxiella burnetii. Infect. Immun. 2002, 70, 6741–6750. [Google Scholar] [CrossRef] [Green Version]
- McCaul, T.F.; Banerjee-Bhatnagar, N.; Williams, J.C. Antigenic differences between Coxiella burnetii cells revealed by postembedding immunoelectron microscopy and immunoblotting. Infect. Immun. 1991, 59, 3243–3253. [Google Scholar] [CrossRef] [Green Version]
- Guevarra, L.A., Jr.; Boado, K.J.O.; Cenidoza, F.B.B.; Imbao, M.; Sia, M.J.G.; Dalmacio, L.M.M. A synthetic peptide analog of in silico-predicted immunogenic epitope unique to dengue virus serotype 2 NS1 antigen specifically binds immunoglobulin G antibodies raised in rabbits. Microbiol. Immunol. 2020, 64, 153–161. [Google Scholar] [CrossRef]
- Kohler, H.; Liebler-Tenorio, E.; Hughes, V.; Stevenson, K.; Bakker, D.; Willemsen, P.; Bay, S.; Ganneau, C.; Biet, F.; Vordermeier, H.M. Interferon-gamma response of mycobacterium avium subsp. Paratuberculosis infected goats to recombinant and synthetic mycobacterial antigens. Front. Vet. Sci. 2021, 8, 645251. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Malik, S.S.; Vergis, J.; Ramanjeneya, S.; Sahu, R.; Pathak, R.; Yadav, J.P.; Dhaka, P.; Barbuddhe, S.B.; Rawool, D.B. Development of the Com1 synthetic peptide-based Latex Agglutination Test (LAT) and its comparative evaluation with commercial indirect-ELISA for sero-screening of coxiellosis in cattle. J. Microbiol. Methods 2019, 162, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Middleton, S.; Steinbach, S.; Coad, M.; McGill, K.; Brady, C.; Duignan, A.; Wiseman, J.; Gormley, E.; Jones, G.J.; Vordermeier, H.M. A molecularly defined skin test reagent for the diagnosis of bovine tuberculosis compatible with vaccination against Johne’s Disease. Sci. Rep. 2021, 11, 2929. [Google Scholar] [CrossRef] [PubMed]
- Polvere, I.; Parrella, A.; Casamassa, G.; D’Andrea, S.; Tizzano, A.; Cardinale, G.; Voccola, S.; Porcaro, P.; Stilo, R.; Vito, P.; et al. Seroprevalence of anti-SARS-CoV-2 IgG and IgM among adults over 65 years old in the south of Italy. Diagnostics 2021, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, S.; Nayak, S.; Williams, T.; di Santa Maria, F.S.; Guedes, M.S.; Chaves, R.C.; Linder, V.; Marques, A.R.; Horn, E.J.; Wong, S.J.; et al. A multiplexed serologic test for diagnosis of lyme disease for point-of-care use. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Murphy, N.; Macchiaverna, N.P.; Cardinal, M.V.; Bhattacharyya, T.; Mertens, P.; Zeippen, N.; Gustin, Y.; Gilleman, Q.; Gurtler, R.E.; Miles, M.A. Lineage-specific rapid diagnostic tests can resolve trypanosoma cruzi TcII/V/VI ecological and epidemiological associations in the argentine chaco. Parasit Vectors 2019, 12, 424. [Google Scholar] [CrossRef] [Green Version]
- Prado, I.C.; Mendes, V.G.; Souza, A.L.A.; Dutra, R.F.; De-Simone, S.G. Electrochemical immunosensor for differential diagnostic of Wuchereria bancrofti using a synthetic peptide. Biosens. Bioelectron. 2018, 113, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Rawool, D.B.; Vinod, V.K.; Malik, S.V.S.; Barbuddhe, S.B. Current approaches for the detection of Coxiella burnetii infection in humans and animals. J. Microbiol. Methods 2020, 179, 106087. [Google Scholar] [CrossRef] [PubMed]
- Noya, O.; Patarroyo, M.E.; Guzman, F.; Alarcon de Noya, B. Immunodiagnosis of parasitic diseases with synthetic peptides. Curr. Protein Pept. Sci. 2003, 4, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Gomara, M.J.; Haro, I. Synthetic peptides for the immunodiagnosis of human diseases. Curr. Med. Chem. 2007, 14, 531–546. [Google Scholar] [CrossRef]
- Kashyap, R.S.; Shekhawat, S.D.; Nayak, A.R.; Purohit, H.J.; Taori, G.M.; Daginawala, H.F. Diagnosis of tuberculosis infection based on synthetic peptides from Mycobacterium tuberculosis antigen 85 complex. Clin. Neurol. Neurosurg. 2013, 115, 678–683. [Google Scholar] [CrossRef]
- Dijkstra, F.; van der Hoek, W.; Wijers, N.; Schimmer, B.; Rietveld, A.; Wijkmans, C.J.; Vellema, P.; Schneeberger, P.M. The 2007–2010 Q fever epidemic in The Netherlands: Characteristics of notified acute Q fever patients and the association with dairy goat farming. FEMS Immunol. Med. Microbiol. 2012, 64, 3–12. [Google Scholar] [CrossRef]
- Schimmer, B.; Notermans, D.W.; Harms, M.G.; Reimerink, J.H.; Bakker, J.; Schneeberger, P.; Mollema, L.; Teunis, P.; van Pelt, W.; van Duynhoven, Y. Low seroprevalence of Q fever in The Netherlands prior to a series of large outbreaks. Epidemiol. Infect. 2012, 140, 27–35. [Google Scholar] [CrossRef]
- Gidding, H.F.; Peng, C.Q.; Graves, S.; Massey, P.D.; Nguyen, C.; Stenos, J.; Quinn, H.E.; McIntyre, P.B.; Durrheim, D.N.; Wood, N. Q fever seroprevalence in Australia suggests one in twenty people have been exposed. Epidemiol. Infect. 2020, 148, e18. [Google Scholar] [CrossRef] [Green Version]
- Anderson, A.D.; Kruszon-Moran, D.; Loftis, A.D.; McQuillan, G.; Nicholson, W.L.; Priestley, R.A.; Candee, A.J.; Patterson, N.E.; Massung, R.F. Seroprevalence of Q fever in the United States, 2003–2004. Am. J. Trop. Med. Hyg. 2009, 81, 691–694. [Google Scholar] [CrossRef]
- Tshokey, T.; Stenos, J.; Durrheim, D.N.; Eastwood, K.; Nguyen, C.; Graves, S.R. Seroprevalence of rickettsial infections and Q fever in Bhutan. PLoS Negl. Trop. Dis. 2017, 11, e0006107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaughey, C.; McKenna, J.; McKenna, C.; Coyle, P.V.; O’Neill, H.J.; Wyatt, D.E.; Smyth, B.; Murray, L.J. Human seroprevalence to Coxiella burnetii (Q fever) in Northern Ireland. Zoonoses Public Health 2008, 55, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Psaroulaki, A.; Hadjichristodoulou, C.; Loukaides, F.; Soteriades, E.; Konstantinidis, A.; Papastergiou, P.; Ioannidou, M.C.; Tselentis, Y. Epidemiological study of Q fever in humans, ruminant animals, and ticks in Cyprus using a geographical information system. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Tapia, T.; Olivares, M.F.; Stenos, J.; Iglesias, R.; Diaz, N.; Vergara, N.; Sotomayor, V.; Gallegos, D.; Soares Magalhaes, R.J.; Acevedo, J.; et al. National seroprevalence of Coxiella burnetii in Chile, 2016–2017. Pathogens 2021, 10, 531. [Google Scholar] [CrossRef]
- Epelboin, L.; Nacher, M.; Mahamat, A.; de Santi, V.P.; Berlioz-Arthaud, A.; Eldin, C.; Abboud, P.; Briolant, S.; Mosnier, E.; Mdo, S.M.G.; et al. Q fever in French guiana: Tip of the iceberg or epidemiological exception? PLoS Negl. Trop. Dis. 2016, 10, e0004598. [Google Scholar] [CrossRef] [Green Version]
- Salarpour, A.; Toroghi, R.; Brujeni, G.N.; Momayez, R. In silico prediction of linear B-cell epitopes for S1 protein of two Iranian 793/B isolates and their changes after 90 serial passaging. Vet. Res. Forum Int. Q. J. 2020, 11, 365–370. [Google Scholar] [CrossRef]
- Moeini, H.; Afridi, S.Q.; Donakonda, S.; Knolle, P.A.; Protzer, U.; Hoffmann, D. Linear B-Cell epitopes in human norovirus GII.4 capsid protein elicit blockade antibodies. Vaccines 2021, 9, 52. [Google Scholar] [CrossRef]
- Sianez-Estrada, L.I.; Rivera-Benitez, J.F.; Rosas-Murrieta, N.H.; Reyes-Leyva, J.; Santos-Lopez, G.; Herrera-Camacho, I. Immunoinformatics approach for predicting epitopes in HN and F proteins of porcine rubulavirus. PLoS ONE 2020, 15, e0239785. [Google Scholar] [CrossRef]
- Conte, F.P.; Tinoco, B.C.; Santos Chaves, T.; Oliveira, R.C.; Figueira Mansur, J.; Mohana-Borges, R.; Lemos, E.R.S.; Neves, P.; Rodrigues-da-Silva, R.N. Identification and validation of specific B-cell epitopes of hantaviruses associated to hemorrhagic fever and renal syndrome. PLoS Negl. Trop. Dis. 2019, 13, e0007915. [Google Scholar] [CrossRef] [Green Version]
- Almeida, M.A.; Almeida-Paes, R.; Guimaraes, A.J.; Valente, R.H.; Soares, C.M.A.; Zancope-Oliveira, R.M. Immunoproteomics reveals pathogen’s antigens involved in homo sapiens-histoplasma capsulatum interaction and specific linear B-cell epitopes in histoplasmosis. Front. Cell. Infect. Microbiol. 2020, 10, 591121. [Google Scholar] [CrossRef]
- Matos, A.D.S.; Rodrigues-da-Silva, R.N.; Soares, I.F.; Baptista, B.O.; de Souza, R.M.; Bitencourt-Chaves, L.; Totino, P.R.R.; Sanchez-Arcila, J.C.; Daniel-Ribeiro, C.T.; Lopez-Camacho, C.; et al. Antibody responses against plasmodium vivax TRAP recombinant and synthetic antigens in naturally exposed individuals from the brazilian amazon. Front. Immunol. 2019, 10, 2230. [Google Scholar] [CrossRef]
- Campos, M.P.; Figueiredo, F.B.; Morgado, F.N.; Renzetti, A.; de Souza, S.M.M.; Pereira, S.A.; Rodrigues-Da-Silva, R.N.; Lima-Junior, J.D.C.; De Luca, P.M. Leishmania infantum virulence factor A2 protein: Linear B-cell epitope mapping and identification of three main linear B-cell epitopes in vaccinated and naturally infected dogs. Front. Immunol. 2018, 9, 1690. [Google Scholar] [CrossRef]
- Zargaran, F.N.; Akya, A.; Rezaeian, S.; Ghadiri, K.; Lorestani, R.C.; Madanchi, H.; Safaei, S.; Rostamian, M. B cell epitopes of four fimbriae antigens of klebsiella pneumoniae: A comprehensive in silico study for vaccine development. Int. J. Pept. Res. Ther. 2021, 27, 875–886. [Google Scholar] [CrossRef]
- Feodorova, V.A.; Lyapina, A.M.; Khizhnyakova, M.A.; Zaitsev, S.S.; Saltykov, Y.V.; Motin, V.L. Yersinia pestis antigen F1 but not LcrV induced humoral and cellular immune responses in humans immunized with live plague vaccine-comparison of immunoinformatic and immunological approaches. Vaccines 2020, 8, 698. [Google Scholar] [CrossRef]
- Tas, S.K.; Kirkik, D.; Ozturk, K.; Tanoglu, A. Determination of B- and T-cell epitopes for Helicobacter pylori cagPAI: An in silico approach. Turk. J. Gastroenterol. 2020, 31, 713–720. [Google Scholar] [CrossRef]
- Jaydari, A.; Forouharmehr, A.; Nazifi, N. Determination of immunodominant scaffolds of Com1 and OmpH antigens of Coxiella burnetii. Microb. Pathog. 2019, 126, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, G.; Duclohier, H.; Thomas, D.; Shechter, E.; Wroblewski, H. Purification and characterization of protein H, the major porin of Pasteurella multocida. J. Bacteriol. 1993, 175, 266–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jameson-Lee, M.; Garduno, R.A.; Hoffman, P.S. DsbA2 (27 kDa Com1-like protein) of legionella pneumophila catalyses extracytoplasmic disulphide-bond formation in proteins including the Dot/Icm type IV secretion system. Mol. Microbiol. 2011, 80, 835–852. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Miura, H.; Ohsumi, K.; Osaki, T.; Taguchi, H.; Yamamoto, T.; Hanawa, T.; Ogata, S.; Kamiya, S. Analysis of the epitopes recognized by mouse monoclonal antibodies directed to Yersinia enterocolitica heat-shock protein 60. Microbiol. Immunol. 1996, 40, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Vizcaino, N.; Zygmunt, M.S.; Verger, J.M.; Grayon, M.; Cloeckaert, A. Localization and characterization of a specific linear epitope of the Brucella DnaK protein. FEMS Microbiol. Lett. 1997, 154, 117–122. [Google Scholar] [CrossRef]
- Wang, W.; Wu, J.; Qiao, J.; Weng, Y.; Zhang, H.; Liao, Q.; Qiu, J.; Chen, C.; Allain, J.P.; Li, C. Evaluation of humoral and cellular immune responses to BP26 and OMP31 epitopes in the attenuated Brucella melitensis vaccinated sheep. Vaccine 2014, 32, 825–833. [Google Scholar] [CrossRef]
- Chen, C.; Bouman, T.J.; Beare, P.A.; Mertens, K.; Zhang, G.Q.; Russell-Lodrigue, K.E.; Hogaboam, J.P.; Peters, B.; Felgner, P.L.; Brown, W.C.; et al. A systematic approach to evaluate humoral and cellular immune responses to Coxiella burnetii immunoreactive antigens. Clin. Microbiol. Infect. 2009, 15 (Suppl. 2), 156–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Psaroulaki, A.; Mathioudaki, E.; Vranakis, I.; Chochlakis, D.; Yachnakis, E.; Kokkini, S.; Xie, H.; Tsiotis, G. In the search of potential serodiagnostic proteins to discriminate between acute and chronic Q fever in humans. Some promising outcomes. Front. Cell. Infect. Microbiol. 2020, 10, 557027. [Google Scholar] [CrossRef] [PubMed]
- Sekeyova, Z.; Kowalczewska, M.; Vincentelli, R.; Decloquement, P.; Flores-Ramirez, G.; Skultety, L.; Raoult, D. Characterization of antigens for Q fever serodiagnostics. Acta Virol. 2010, 54, 173–180. [Google Scholar] [CrossRef]
- Dangel, L.; Koempf, D.; Fischer, S.F. Comparison of different commercially available enzyme-linked immunosorbent assays with immunofluorescence test for detection of phase II IgG and IgM antibodies to Coxiella burnetii. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Herremans, T.; Hogema, B.M.; Nabuurs, M.; Peeters, M.; Wegdam-Blans, M.; Schneeberger, P.; Nijhuis, C.; Notermans, D.W.; Galama, J.; Horrevorts, A.; et al. Comparison of the performance of IFA, CFA, and ELISA assays for the serodiagnosis of acute Q fever by quality assessment. Diagn. Microbiol. Infect. Dis. 2013, 75, 16–21. [Google Scholar] [CrossRef]
- Martinez, E.; Cantet, F.; Fava, L.; Norville, I.; Bonazzi, M. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog. 2014, 10, e1004013. [Google Scholar] [CrossRef] [Green Version]
- Seshu, J.; McIvor, K.L.; Mallavia, L.P. Antibodies are generated during infection to Coxiella burnetii macrophage infectivity potentiator protein (Cb-Mip). Microbiol. Immunol. 1997, 41, 371–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues-da-Silva, R.N.; Soares, I.F.; Lopez-Camacho, C.; da Silva, J.H.M.; Perce-da-Silva, D.S.; Teva, A.; Franco, A.M.R.; Pinheiro, F.G.; Chaves, L.B.; Pratt-Riccio, L.R.; et al. Plasmodium vivax Cell-traversal protein for ookinetes and sporozoites: Naturally acquired humoral immune response and b-cell epitope mapping in brazilian amazon inhabitants. Front. Immunol. 2017, 8, 77. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Niu, D.; Wen, B.; Chen, M.; Qiu, L.; Zhang, J. Protective immunity against Q fever induced with a recombinant P1 antigen fused with HspB of Coxiella burnetii. Ann. N. Y. Acad. Sci. 2005, 1063, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Scholzen, A.; Richard, G.; Moise, L.; Baeten, L.A.; Reeves, P.M.; Martin, W.D.; Brauns, T.A.; Boyle, C.M.; Paul, S.R.; Bucala, R.; et al. Promiscuous Coxiella burnetii CD4 epitope clusters associated with human recall responses are candidates for a novel T-cell targeted multi-epitope Q fever vaccine. Front. Immunol. 2019, 10, 207. [Google Scholar] [CrossRef]
- Prado, I.C.; Chino, M.; Santos, A.L.D.; Souza, A.L.A.; Pinho, L.G.; Lemos, E.R.S.; De-Simone, S.G. Development of an electrochemical immunosensor for the diagnostic testing of spotted fever using synthetic peptides. Biosens. Bioelectron. 2018, 100, 115–121. [Google Scholar] [CrossRef]
- Larsen, J.E.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emini, E.A.; Hughes, J.V.; Perlow, D.S.; Boger, J. Induction of hepatitis a virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Raghava, G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins 2006, 65, 40–48. [Google Scholar] [CrossRef]
- Zaharieva, N.; Dimitrov, I.; Flower, D.R.; Doytchinova, I. VaxiJen dataset of bacterial immunogens: An update. Curr. Comput. Aided Drug Des. 2019, 15, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Kim, D.E.; Ovchinnikov, S.; Baker, D.; DiMaio, F. Automatic structure prediction of oligomeric assemblies using Robetta in CASP12. Proteins 2018, 86 (Suppl. 1), 283–291. [Google Scholar] [CrossRef] [PubMed]
- Chivian, D.; Kim, D.E.; Malmstrom, L.; Schonbrun, J.; Rohl, C.A.; Baker, D. Prediction of CASP6 structures using automated Robetta protocols. Proteins 2005, 61 (Suppl. 7), 157–166. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chivian, D.; Kim, D.E.; Malmstrom, L.; Bradley, P.; Robertson, T.; Murphy, P.; Strauss, C.E.; Bonneau, R.; Rohl, C.A.; Baker, D. Automated prediction of CASP-5 structures using the Robetta server. Proteins 2003, 53 (Suppl. 6), 524–533. [Google Scholar] [CrossRef]
- Gierut, A.M.; Dabrowski-Tumanski, P.; Niemyska, W.; Millett, K.C.; Sulkowska, J.I. PyLink: A PyMOL plugin to identify links. Bioinformatics 2019, 35, 3166–3168. [Google Scholar] [CrossRef] [PubMed]
- Grell, L.; Parkin, C.; Slatest, L.; Craig, P.A. EZ-Viz, a tool for simplifying molecular viewing in PyMOL. BioChem. Mol. Biol. Educ. 2006, 34, 402–407. [Google Scholar] [CrossRef]
- Baek, M.; Park, T.; Heo, L.; Seok, C. Modeling protein homo-oligomer structures with GalaxyHomomer web server. Methods Mol. Biol 2020, 2165, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; Park, T.; Heo, L.; Park, C.; Seok, C. GalaxyHomomer: A web server for protein homo-oligomer structure prediction from a monomer sequence or structure. Nucleic Acids Res. 2017, 45, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Dupont, H.T.; Thirion, X.; Raoult, D. Q fever serology: Cutoff determination for microimmunofluorescence. Clin. Diagn. Lab. Immunol. 1994, 1, 189–196. [Google Scholar] [CrossRef] [PubMed]
| Protein | Sequence | Bepipred | ABCpred | ESA |
|---|---|---|---|---|
| OMP-H | 51-QFSPQREKM-59 | × | × | × |
| 91-EIQNDESTLRQQQQQF-106 | × | × | × | |
| Com-1 | 26-FSFSPQQVK-34 | × | × | - |
| 57-ALQKKTEAQQEEHAQQAIKE-76 | × | × | × | |
| 83-FNDPASPVAGNPHGN-96 | × | × | - | |
| 191-KKDMDNPAIQKQLRDN-206 | × | × | × | |
| OMP-P1 | 43-GYKSYTYDQ-51 | × | - | × |
| 98-KAQYQYDNV-106 | - | × | × | |
| 197-LSYDYALYRSKSN-209 | × | × | ||
| 215-SATASAEGTAIG-226 | × | × |
| Protein | Epitope | Length (mers) | Vaxijen Score |
|---|---|---|---|
| OMP-H | OMP-H(51–59) | 9 | 0.524 |
| OMP-H(91–106) | 16 | 0.671 | |
| Com-1 | Com-1(26–34) | 9 | 1.435 |
| Com-1(57–76) | 20 | 0.923 | |
| Com-1(83–96) | 14 | 0.139 | |
| Com-1(191–206) | 16 | 0.418 | |
| OMP-P1 | OMP-P1(43–51) | 9 | 0.383 |
| OMP-P1(98–106) | 9 | 0.598 | |
| OMP-P1(197–209) | 13 | 0.598 | |
| OMP-P1(215–227) | 13 | 1.359 |
| Overall (n = 57) | C. burnetii Seroreactive (n = 26) | C. burnetii Non-Reactive (n = 31) | |
|---|---|---|---|
| Age-Median (IR) | 33 (22–49) | 33 (21–53) | 33 (23–48) |
| Gender, n (%) | |||
| Male | 32 (66%) | 17 (65%) | 15 (48%) |
| Female | 25 (42%) | 9 (35%) | 16 (52%) |
| Symptomatology, n (%) | |||
| Fever | 24 (42%) | 10 (38%) | 14 (45%) |
| Nausea | 7 (12%) | 4 (15%) | 3 (10%) |
| Endocarditis | 6 (11%) | 4 (15%) | 2 (6%) |
| Hemorrhagic manifestations | 1 (2%) | 0 | 1 (3%) |
| Myalgia | 13 (23%) | 4 (15%) | 9 (29%) |
| Respiratory manifestations | 11 (19%) | 3 (12%) | 8 (26%) |
| Prostration | 18 (32%) | 6 (23%) | 12 (39%) |
| Group | ID | OMP-H(51–59) | OMP-H(91–106) | Com-1(57–76) | Com-1(191–206) | OMP-P1(197–209) | OMP-P1(215–227) |
|---|---|---|---|---|---|---|---|
| C. burnetiiseroreactive patients | 318/12 | 0.827 | 0.671 | 0.813 | 0.920 | 0.765 | 0.912 |
| 487/12 | 1.006 | 0.736 | 1.020 | 0.856 | 0.610 | 0.869 | |
| 69/13 | 0.822 | 0.736 | 0.593 | 0.583 | 0.869 | 0.856 | |
| 207/17 | 0.651 | 0.891 | 0.890 | 0.910 | 0.648 | 1.060 | |
| 464/16 | 0.791 | 0.744 | 0.824 | 0.774 | 0.663 | 0.668 | |
| 466/16 | 1.038 | 1.279 | 0.271 | 1.089 | 0.471 | 0.931 | |
| 471/16 | 1.094 | 1.042 | 0.946 | 1.045 | 1.418 | 1.259 | |
| 104/12 | 1.430 | 1.634 | 1.916 | 1.887 | 1.420 | 1.695 | |
| 551/12 | 1.816 | 1.601 | 1.318 | 0.716 | 0.849 | 1.625 | |
| 118/13 | 2.265 | 1.260 | 1.408 | 1.242 | 1.585 | 1.224 | |
| 463/16 | 1.100 | 0.826 | 1.001 | 3.540 | 0.892 | 1.014 | |
| 126/13 | 1.250 | 1.475 | 0.611 | 0.271 | 0.689 | 0.941 | |
| 134/13 | 0.997 | 0.882 | 0.837 | 0.296 | 0.740 | 1.757 | |
| 130/16 | 2.075 | 2.006 | 1.712 | 1.180 | 0.824 | 2..014 | |
| 140/13 | 1.137 | 0.193 | 1.132 | 1.060 | 0.915 | 0.901 | |
| 151/13 | 1.055 | 0.180 | 1.137 | 1.298 | 0.921 | 0.877 | |
| 204/13 | 1.250 | 0.192 | 1.319 | 1.197 | 0.796 | 1.141 | |
| 253/13 | 0.981 | 1.744 | 1.340 | 1.864 | 0.993 | 2.038 | |
| 264/13 | 3.086 | 4.270 | 4.916 | 6.404 | 0.996 | 4.994 | |
| 304/13 | 1.369 | 1.808 | 1.716 | 2.371 | 0.990 | 1.885 | |
| 81/14 | 0.432 | 0.498 | 0.589 | 0.462 | 0.814 | 0.663 | |
| 20/16 | 0.835 | 0.550 | 0.664 | 0.606 | 0.658 | 0.818 | |
| 480/16 | 0.412 | 0.640 | 0.552 | 540 | 0.110 | 0.628 | |
| 490/17 | 2.072 | 2.088 | 2.215 | 2006 | 1.338 | 1.940 | |
| 239/18 | 1.469 | 1.835 | 1.948 | 1676 | 1.216 | 1.604 | |
| 311/18 | 2.042 | 2.497 | 2.372 | 2256 | 1.379 | 2.175 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontes, S.d.S.; Maia, F.d.M.; Ataides, L.S.; Conte, F.P.; Lima-Junior, J.d.C.; Rozental, T.; da Silva Assis, M.R.; Júnior, A.A.P.; Fernandes, J.; de Lemos, E.R.S.; et al. Identification of Immunogenic Linear B-Cell Epitopes in C. burnetii Outer Membrane Proteins Using Immunoinformatics Approaches Reveals Potential Targets of Persistent Infections. Pathogens 2021, 10, 1250. https://doi.org/10.3390/pathogens10101250
Fontes SdS, Maia FdM, Ataides LS, Conte FP, Lima-Junior JdC, Rozental T, da Silva Assis MR, Júnior AAP, Fernandes J, de Lemos ERS, et al. Identification of Immunogenic Linear B-Cell Epitopes in C. burnetii Outer Membrane Proteins Using Immunoinformatics Approaches Reveals Potential Targets of Persistent Infections. Pathogens. 2021; 10(10):1250. https://doi.org/10.3390/pathogens10101250
Chicago/Turabian StyleFontes, Sílvia da Silva, Fernanda de Moraes Maia, Laura Santa’Anna Ataides, Fernando Paiva Conte, Josué da Costa Lima-Junior, Tatiana Rozental, Matheus Ribeiro da Silva Assis, Adonai Alvino Pessoa Júnior, Jorlan Fernandes, Elba Regina Sampaio de Lemos, and et al. 2021. "Identification of Immunogenic Linear B-Cell Epitopes in C. burnetii Outer Membrane Proteins Using Immunoinformatics Approaches Reveals Potential Targets of Persistent Infections" Pathogens 10, no. 10: 1250. https://doi.org/10.3390/pathogens10101250
APA StyleFontes, S. d. S., Maia, F. d. M., Ataides, L. S., Conte, F. P., Lima-Junior, J. d. C., Rozental, T., da Silva Assis, M. R., Júnior, A. A. P., Fernandes, J., de Lemos, E. R. S., & Rodrigues-da-Silva, R. N. (2021). Identification of Immunogenic Linear B-Cell Epitopes in C. burnetii Outer Membrane Proteins Using Immunoinformatics Approaches Reveals Potential Targets of Persistent Infections. Pathogens, 10(10), 1250. https://doi.org/10.3390/pathogens10101250