Beyond the IFA: Revisiting the ELISA as a More Sensitive, Objective, and Quantitative Evaluation of Spotted Fever Group Rickettsia Exposure
Abstract
1. Introduction
2. Results and Discussion
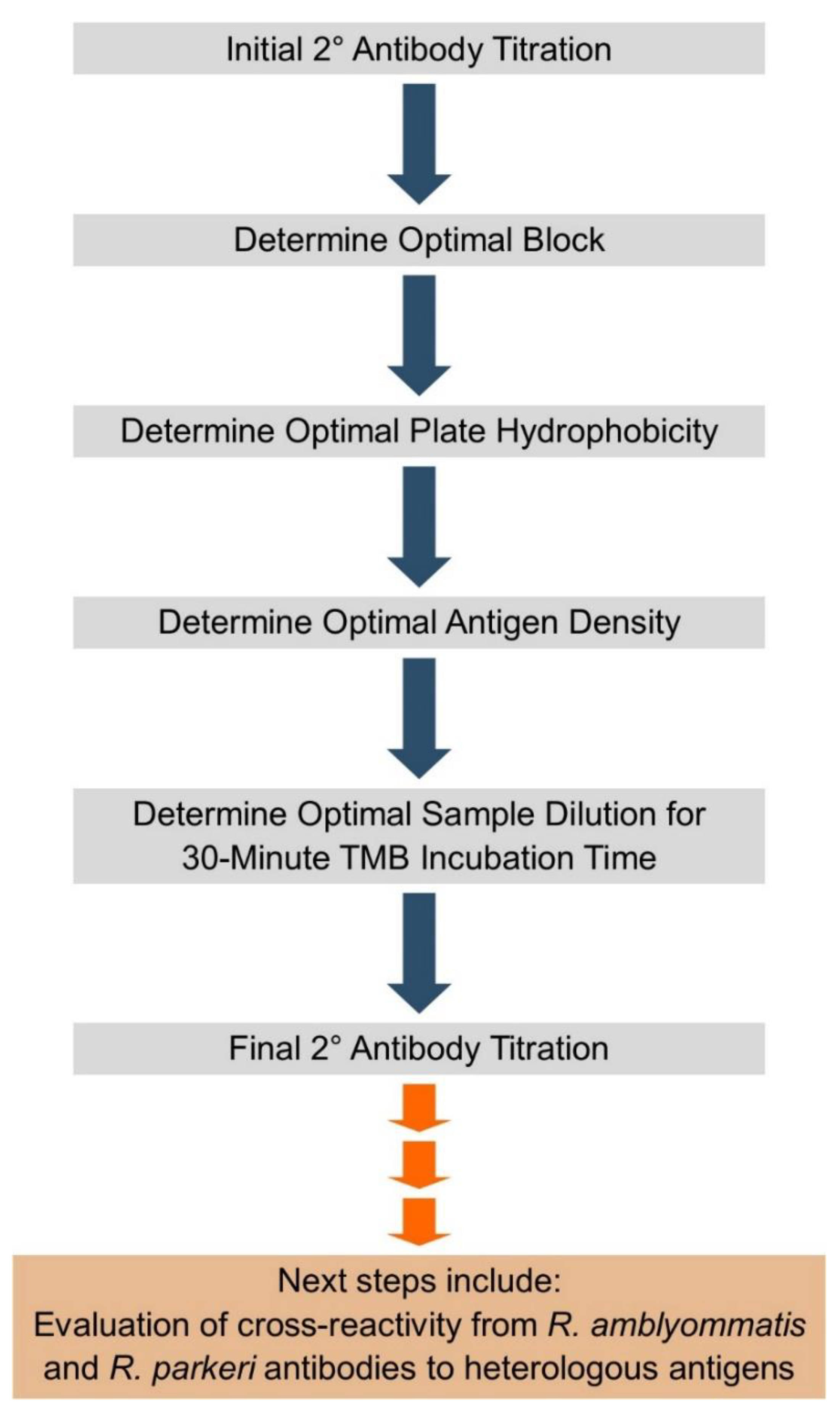
3. Materials and Methods
3.1. Sample Collection
3.2. Antigen Preparation and Plating
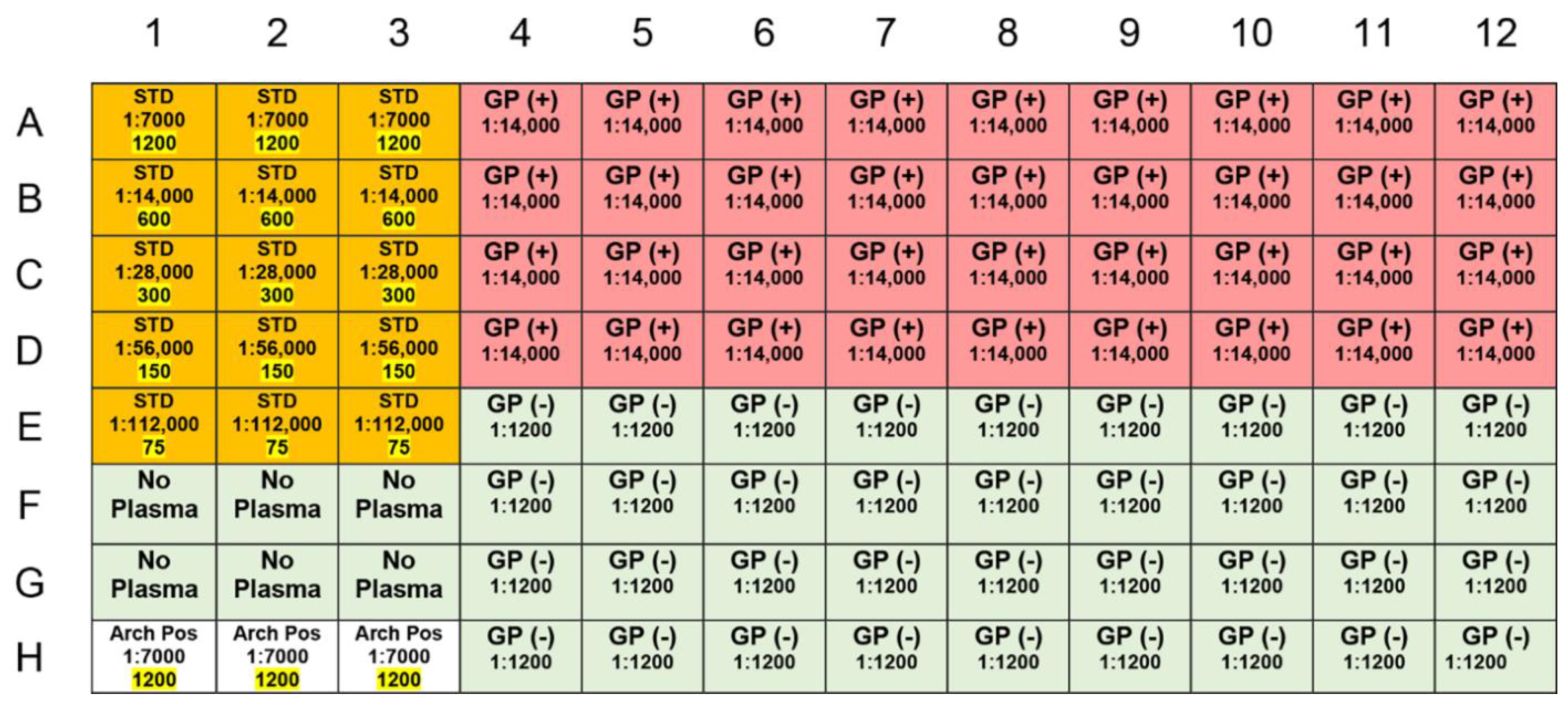
3.3. Assay
3.4. Data Acquisition and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.E.; et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef] [PubMed]
- Blanton, L.S. The Rickettsioses: A practical update. Infect. Dis. Clin. N. Am. 2019, 33, 213–229. [Google Scholar] [CrossRef]
- Paddock, C.D.; Sumner, J.W.; Comer, J.A.; Zaki, S.R.; Goldsmith, C.S.; Goddard, J.; McLellan, S.L.; Tamminga, C.L.; Ohl, C.A. Rickettsia parkeri: A newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 2004, 38, 805–811. [Google Scholar] [CrossRef]
- Sumner, J.W.; Durden, L.A.; Goddard, J.; Stromdahl, E.Y.; Clark, K.L.; Reeves, W.K.; Paddock, C.D. Gulf Coast ticks (Amblyomma maculatum) and Rickettsia parkeri, United States. Emerg. Infect. Dis. 2007, 13, 751–753. [Google Scholar] [CrossRef]
- Yaglom, H.D.; Casal, M.; Carson, S.; O’Grady, C.L.; Dominguez, V.; Singleton, J.; Chung, I.; Lodge, H.; Paddock, C.D. Expanding recognition of Rickettsia parkeri rickettsiosis in southern Arizona, 2016–2017. Vector Borne Zoonotic Dis. 2019, 20, 82–87. [Google Scholar] [CrossRef]
- Shapiro, M.R.; Fritz, C.L.; Tait, K.; Paddock, C.D.; Nicholson, W.L.; Abramowicz, K.F.; Karpathy, S.E.; Dasch, G.A.; Sumner, J.W.; Adem, P.V.; et al. Rickettsia 364D: A newly recognized cause of eschar-associated illness in California. Clin. Infect. Dis. 2010, 50, 541–548. [Google Scholar] [CrossRef]
- Karpathy, S.E.; Slater, K.S.; Goldsmith, C.S.; Nicholson, W.L.; Paddock, C.D. Rickettsia amblyommatis sp. nov., a spotted fever group Rickettsia associated with multiple species of Amblyomma ticks in North, Central and South America. Int. J. Syst. Evol. Microbiol. 2016, 66, 5236–5243. [Google Scholar] [CrossRef]
- Goddard, J.; Varela-Stokes, A.S. Role of the lone star tick, Amblyomma americanum (L.), in human and animal diseases. Vet. Parasitol. 2009, 160, 1–12. [Google Scholar] [CrossRef]
- Marshall, G.S.; Stout, G.G.; Jacobs, R.F.; Schutze, G.E.; Paxton, H.; Buckingham, S.C.; DeVincenzo, J.P.; Jackson, M.A.; San Joaquin, V.H.; Standaert, S.M.; et al. Antibodies reactive to Rickettsia rickettsii among children living in the southeast and south central regions of the United States. JAMA Pediatr. 2003, 157, 443–448. [Google Scholar] [CrossRef]
- Vaughn, M.F.; Delisle, J.; Johnson, J.; Daves, G.; Williams, C.; Reber, J.; Mendell, N.L.; Bouyer, D.H.; Nicholson, W.L.; Moncayo, A.C.; et al. Seroepidemiologic study of human infections with spotted fever group Rickettsiae in North Carolina. J. Clin. Microbiol. 2014, 52, 3960–3966. [Google Scholar] [CrossRef]
- Graf, P.C.; Chretien, J.P.; Ung, L.; Gaydos, J.C.; Richards, A.L. Prevalence of seropositivity to spotted fever group rickettsiae and Anaplasma phagocytophilum in a large, demographically diverse US sample. Clin. Infect. Dis. 2008, 46, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Delisle, J.; Mendell, N.L.; Stull-Lane, A.; Bloch, K.C.; Bouyer, D.H.; Moncayo, A.C. Human infections by multiple spotted fever group rickettsiae in Tennessee. Am. J. Trop. Med. Hyg. 2016, 94, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Ricketts, H.T. The transmission of Rocky Mountain spotted fever by the bite of the wood-tick (Dermacentor occidentalis). JAMA 1906, XLVII, 358. [Google Scholar] [CrossRef]
- Wolbach, S.B. The etiology of Rocky Mountain spotted fever: (A Preliminary report.). J. Med. Res. 1916, 34, 121–126.1. [Google Scholar] [PubMed]
- Doyle, A.; McGarry, M.P.; Lee, N.A.; Lee, J.J. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 2012, 21, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Birck, M.M.; Tveden-Nyborg, P.; Lindblad, M.M.; Lykkesfeldt, J. Non-terminal blood sampling techniques in guinea pigs. J. Vis. Exp. 2014. [Google Scholar] [CrossRef] [PubMed]
- Goddard, J. Experimental infection of lone star ticks, Amblyomma americanum (L.), with Rickettsia parkeri and exposure of guinea pigs to the agent. J. Med. Entomol. 2003, 40, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Stanley, H.M.; Ford, S.L.; Snellgrove, A.N.; Hartzer, K.; Smith, E.B.; Krapiunaya, I.; Levin, M.L. The ability of the invasive Asian Longhorned Tick Haemaphysalis longicornis (Acari: Ixodidae) to acquire and transmit Rickettsia rickettsii (Rickettsiales: Rickettsiaceae), the agent of Rocky Mountain spotted fever, under laboratory conditions. J. Med. Entomol. 2020, 57, 1635–1639. [Google Scholar] [CrossRef]
- Paddock, C.D.; Finley, R.W.; Wright, C.S.; Robinson, H.N.; Schrodt, B.J.; Lane, C.C.; Ekenna, O.; Blass, M.A.; Tamminga, C.L.; Ohl, C.A.; et al. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin. Infect. Dis. 2008, 47, 1188–1196. [Google Scholar] [CrossRef]
- Spotted Fever Rickettsiosis (Including Rocky Mountain Spotted Fever) (SFR, Including RMSF) 2020 Case Definition. Center for Disease Control and Prevention, Atlanta, GA. Available online: https://wwwn.cdc.gov/nndss/conditions/spotted-fever-rickettsiosis/case-definition/2020/ (accessed on 4 December 2020).
- Clements, M.L.; Dumler, J.S.; Fiset, P.; Wisseman, C.L., Jr.; Snyder, M.J.; Levine, M.M. Serodiagnosis of Rocky Mountain spotted fever: Comparison of IgM and IgG enzyme-linked immunosorbent assays and indirect fluorescent antibody test. J. Infect. Dis. 1983, 148, 876–880. [Google Scholar] [CrossRef]
- Keysary, A.; Strenger, C. Use of enzyme-linked immunosorbent assay techniques with cross-reacting human sera in diagnosis of murine typhus and spotted fever. J. Clin. Microbiol. 1997, 35, 1034–1035. [Google Scholar] [CrossRef] [PubMed]
- Lokida, D.; Sudarmono, P.; Kosasih, H.; Butar-Butar, D.P.; Salim, G.; Antonjaya, U.; Sari, R.A.; Aman, A.T.; Parwati, I.; Arif, M.; et al. Comparison of commercial enzyme-linked immunosorbent assay and immunofluorescence assay for diagnosis of acute Rickettsia typhi infections. Vector Borne Zoonotic Dis. 2020, 20, 93–99. [Google Scholar] [CrossRef]
- Land, M.V.; Ching, W.M.; Dasch, G.A.; Zhang, Z.; Kelly, D.J.; Graves, S.R.; Devine, P.L. Evaluation of a commercially available recombinant-protein enzyme-linked immunosorbent assay for detection of antibodies produced in scrub typhus rickettsial infections. J. Clin. Microbiol. 2000, 38, 2701–2705. [Google Scholar] [CrossRef] [PubMed]
- Munderloh, U.G.; Liu, Y.; Wang, M.; Chen, C.; Kurtti, T.J. Establishment, maintenance and description of cell lines from the tick Ixodes scapularis. J. Parasitol. 1994, 80, 533–553. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alugubelly, N.; Stokes, J.V.; Cross, C.E.; Ross, A.-M.L.; Crawford, A.E.; Fiihr, G.F.; Varela-Stokes, A.S. Beyond the IFA: Revisiting the ELISA as a More Sensitive, Objective, and Quantitative Evaluation of Spotted Fever Group Rickettsia Exposure. Pathogens 2021, 10, 88. https://doi.org/10.3390/pathogens10020088
Alugubelly N, Stokes JV, Cross CE, Ross A-ML, Crawford AE, Fiihr GF, Varela-Stokes AS. Beyond the IFA: Revisiting the ELISA as a More Sensitive, Objective, and Quantitative Evaluation of Spotted Fever Group Rickettsia Exposure. Pathogens. 2021; 10(2):88. https://doi.org/10.3390/pathogens10020088
Chicago/Turabian StyleAlugubelly, Navatha, John V. Stokes, Claire E. Cross, Anne-Marie L. Ross, Anna E. Crawford, Gabrielle F. Fiihr, and Andrea S. Varela-Stokes. 2021. "Beyond the IFA: Revisiting the ELISA as a More Sensitive, Objective, and Quantitative Evaluation of Spotted Fever Group Rickettsia Exposure" Pathogens 10, no. 2: 88. https://doi.org/10.3390/pathogens10020088
APA StyleAlugubelly, N., Stokes, J. V., Cross, C. E., Ross, A.-M. L., Crawford, A. E., Fiihr, G. F., & Varela-Stokes, A. S. (2021). Beyond the IFA: Revisiting the ELISA as a More Sensitive, Objective, and Quantitative Evaluation of Spotted Fever Group Rickettsia Exposure. Pathogens, 10(2), 88. https://doi.org/10.3390/pathogens10020088






