Skin in the Game: An Assay to Monitor Leukocyte Infiltration in Dermal Lesions of a Guinea Pig Model for Tick-Borne Rickettsiosis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Tissue Dissociation Optimization
2.2. Antibody Titrations
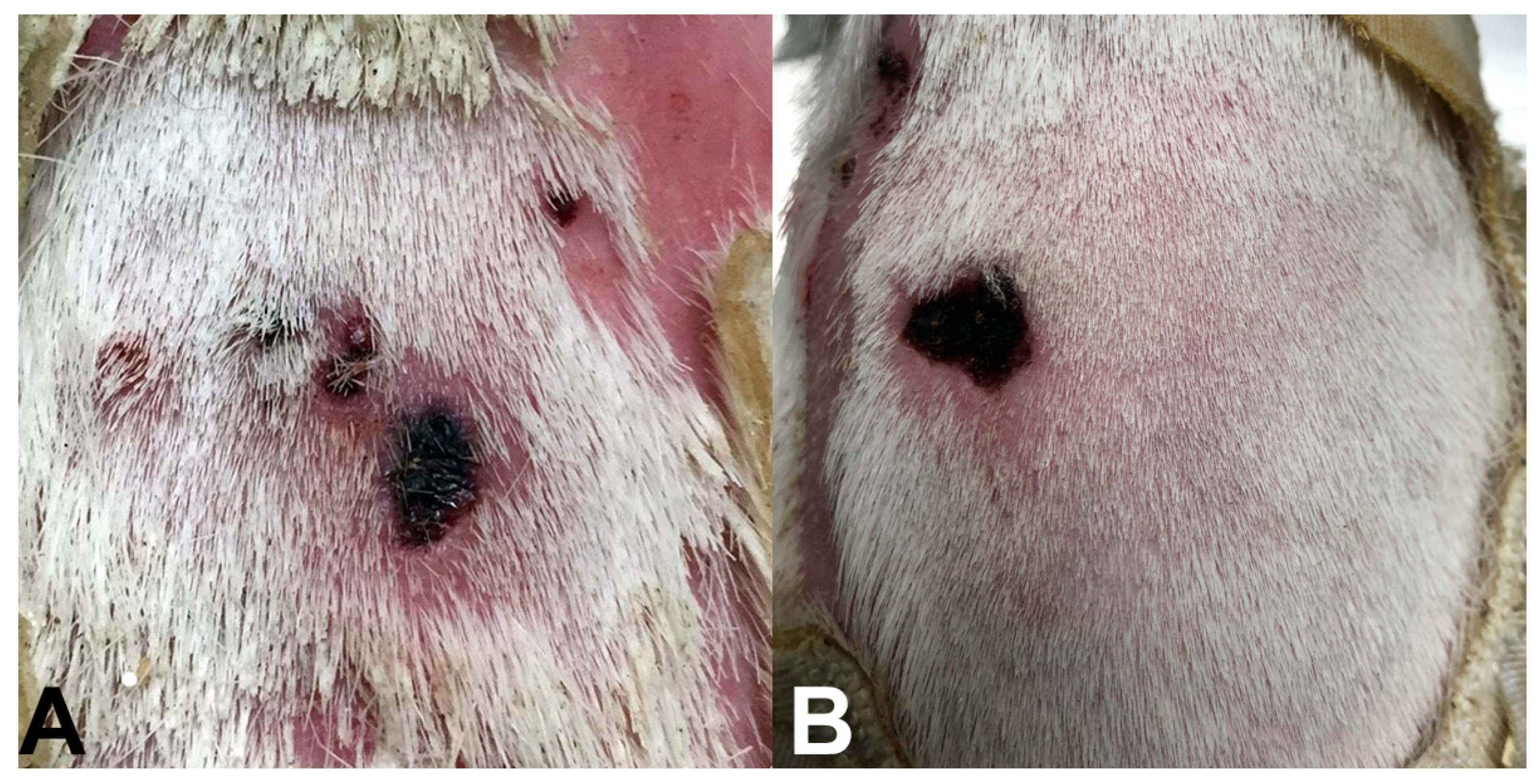
2.3. Leukocyte Infiltration Assays
2.4. Limitations
3. Materials and Methods
3.1. Sample Collection
3.2. Panel Design
3.3. Tissue Dissociation
3.4. Immunofluorescent Staining
3.5. Data Acquisition and Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matejuk, A. Skin Immunity. Arch. Immunol. Ther. Exp. (Warsz) 2018, 66, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.V.; Soulika, A.M. The dynamics of the skin’s immune system. Int. J. Mol. Sci. 2019, 20, 1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Röltgen, K.; Pluschke, G. Buruli ulcer: The efficacy of innate immune defense may be a key determinant for the outcome of infection with Mycobacterium ulcerans. Front. Microbiol. 2020, 11, 1018. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, R.; Lindsey, N.P.; Fischer, M.; Gregory, C.J.; Hinckley, A.F.; Mead, P.S.; Paz-Bailey, G.; Waterman, S.H.; Drexler, N.A.; Kersh, G.J.; et al. Vital Signs: Trends in reported vectorborne disease cases-United States and Territories, 2004–2016. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 496–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wikel, S. Ticks and tick-borne pathogens at the cutaneous interface: Host defenses, tick countermeasures, and a suitable environment for pathogen establishment. Front. Microbiol. 2013, 4, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stratford, A.F.; Zoutman, D.E.; Davidson, J.S. Effect of lidocaine and epinephrine on Staphylococcus aureus in a guinea pig model of surgical wound infection. Plast. Reconstr. Surg. 2002, 110, 1275–1279. [Google Scholar]
- Tsouh, P.V.; Addo, P.; Yeboah-Manu, D.; Boyom, F.F. Methods used in preclinical assessment of anti-Buruli ulcer agents: A global perspective. J. Pharmacol. Toxicol. Methods 2015, 73, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Stokes, J.V.; Walker, D.H.; Varela-Stokes, A.S. The guinea pig model for tick-borne spotted fever rickettsioses: A second look. Ticks Tick Borne Dis. 2020, 110, 101538. [Google Scholar] [CrossRef]
- Grasperge, B.J.; Reif, K.E.; Morgan, T.D.; Sunyakumthorn, P.; Bynog, J.; Paddock, C.D.; Macaluso, K.R. Susceptibility of inbred mice to Rickettsia parkeri. Infect. Immun. 2012, 80, 1846–1852. [Google Scholar] [CrossRef] [Green Version]
- Grasperge, B.J.; Morgan, T.W.; Paddock, C.D.; Peterson, K.E.; Macaluso, K.R. Feeding by Amblyomma maculatum (Acari: Ixodidae) enhances Rickettsia parkeri (Rickettsiales: Rickettsiaceae) infection in the skin. J. Med. Entomol. 2014, 51, 855–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banajee, K.H.; Verhoeve, V.I.; Harris, E.K.; Macaluso, K.R. Effect of Amblyomma maculatum (Acari: Ixodidae) saliva on the acute cutaneous immune response to Rickettsia parkeri infection in a murine model. J. Med. Entomol. 2016, 53, 1252–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padilla-Carlin, D.J.; McMurray, D.N.; Hickey, A.J. The guinea pig as a model of infectious diseases. Comp. Med. 2008, 58, 324–340. [Google Scholar] [PubMed]
- Saito, T.B.; Bechelli, J.; Smalley, C.; Karim, S.; Walker, D.H. Vector tick transmission model of spotted fever rickettsiosis. Am. J. Pathol. 2019, 189, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Couzin-Frankel, J. When mice mislead. Science 2013, 342, 922–925. [Google Scholar] [CrossRef]
- Bechah, Y.; Socolovschi, C.; Raoult, D. Identification of rickettsial infections by using cutaneous swab specimens and PCR. Emerg. Infect. Dis. 2011, 17, 83–86. [Google Scholar] [CrossRef]
- Myers, T.; Lalana, T.; Dent, M.; Jiang, J.; Daly, P.L.; Maguire, J.D.; Rickards, A.L. Detecting Rickettsia parkeri infection from eschar swab specimens. Emerg. Infect. Dis. 2013, 19, 778–780. [Google Scholar] [CrossRef]
- Paddock, C.D.; Sumner, J.W.; Comer, J.A.; Zaki, S.R.; Goldsmith, C.S.; Goddard, J.; McLellan, S.L.F.; Tamminga, C.L.; Ohl, C.A. Rickettsia parkeri: A newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 2004, 38, 805–811. [Google Scholar] [CrossRef] [Green Version]
- Oakley, O.R.; Kim, H.; El-Amouri, I.; Lin, P.C.; Cho, J.; Bani-Ahmad, M.; Ko, C. Periovulatory leukocyte infiltration in the rat ovary. Endocrinology 2010, 151, 4551–4559. [Google Scholar] [CrossRef] [Green Version]
- Kristof, M.N.; Allen, P.E.; Yutzy, L.D.; Thibodaux, B.; Paddock, C.D.; Martinez, J.J. Significant growth by Rickettsia species within human macrophage-like cells is a phenotype correlated with the ability to cause disease in mammals. Pathogens 2021, 10, 228. [Google Scholar] [CrossRef]
- Arroyave, E.; Hyseni, I.; Burkhardt, N.; Kuo, Y.-F.; Wang, T.; Munderloh, U.; Fang, R. Rickettsia parkeri with a genetically disrupted phage integrase gene exhibits attenuated virulence and induces protective immunity against fatal rickettsioses in mice. Pathogens 2021, 10, 819. [Google Scholar] [CrossRef] [PubMed]
- Paddock, C.D.; Finley, R.W.; Wright, C.S.; Robinson, H.N.; Schrodt, B.J.; Lane, C.C.; Ekenna, O.; Blass, M.A.; Tamminga, C.L.; Ohl, C.A.; et al. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin. Infect. Dis. 2008, 47, 1188–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrick, K.L.; Pena, S.A.; Yaglom, H.D.; Layton, B.J.; Moors, A.; Loftis, A.D.; Condit, M.E.; Singleton, J.; Kato, C.Y.; Denison, A.M.; et al. Rickettsia parkeri Rickettsiosis, Arizona, USA. Emerg. Infect. Dis. 2016, 22, 780–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anacker, R.L.; Mann, R.E.; Gonzales, C. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J. Clin. Microbiol. 1987, 25, 167–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard, J. Experimental infection of lone star ticks, Amblyomma americanum (L.), with Rickettsia parkeri and exposure of guinea pigs to the agent. J. Med. Entomol. 2003, 110, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.V.; Crawford, A.E.; Cross, C.E.; Ross, A.L.; Walker, J.D.; Willeford, B.V.; Varela-Stokes, A.S. An optimized five-color/seven-parameter flow cytometry panel for immunophenotyping guinea pig peripheral blood lymphocytes. J. Immunol Methods 2020, 476, 112682. [Google Scholar] [CrossRef] [PubMed]
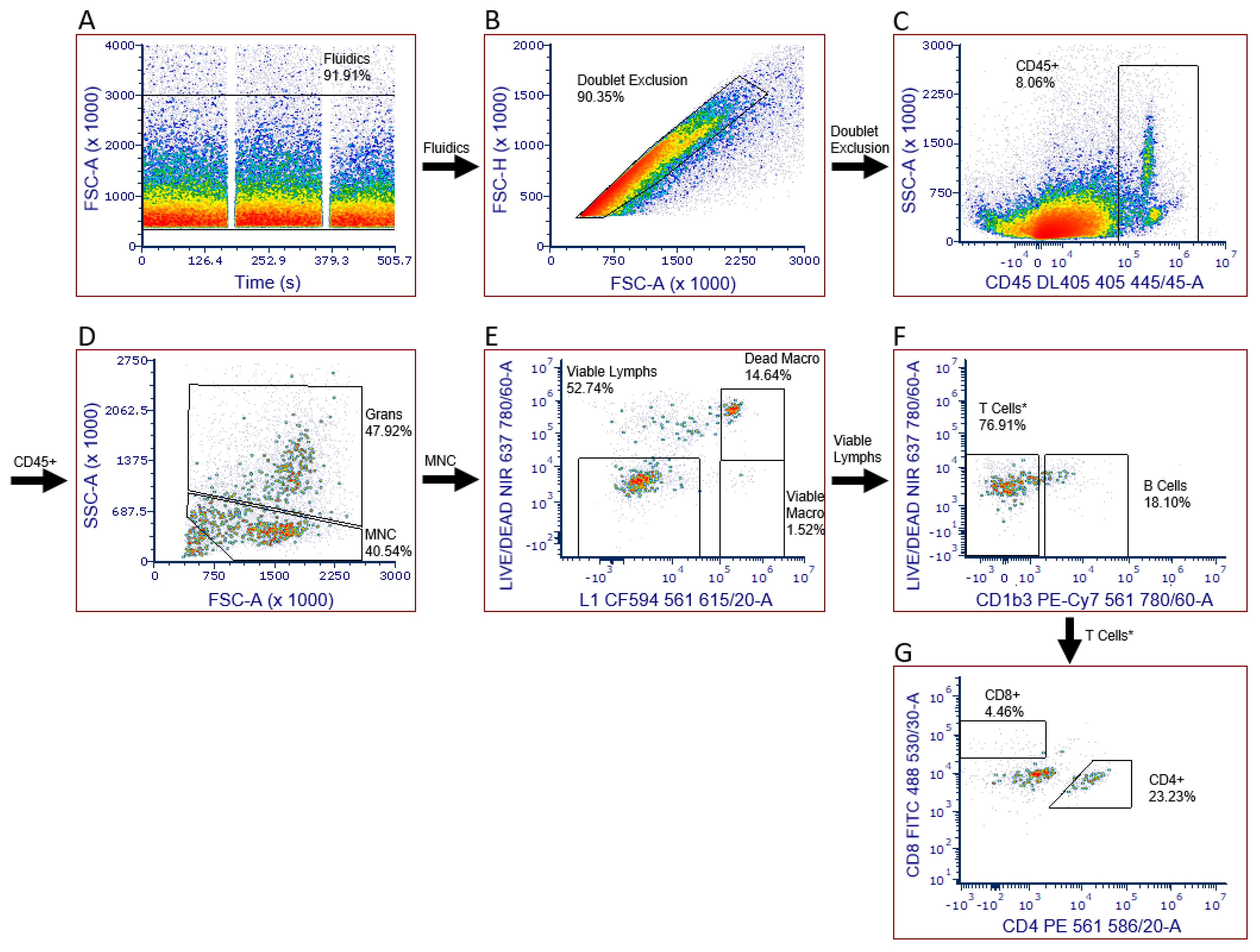
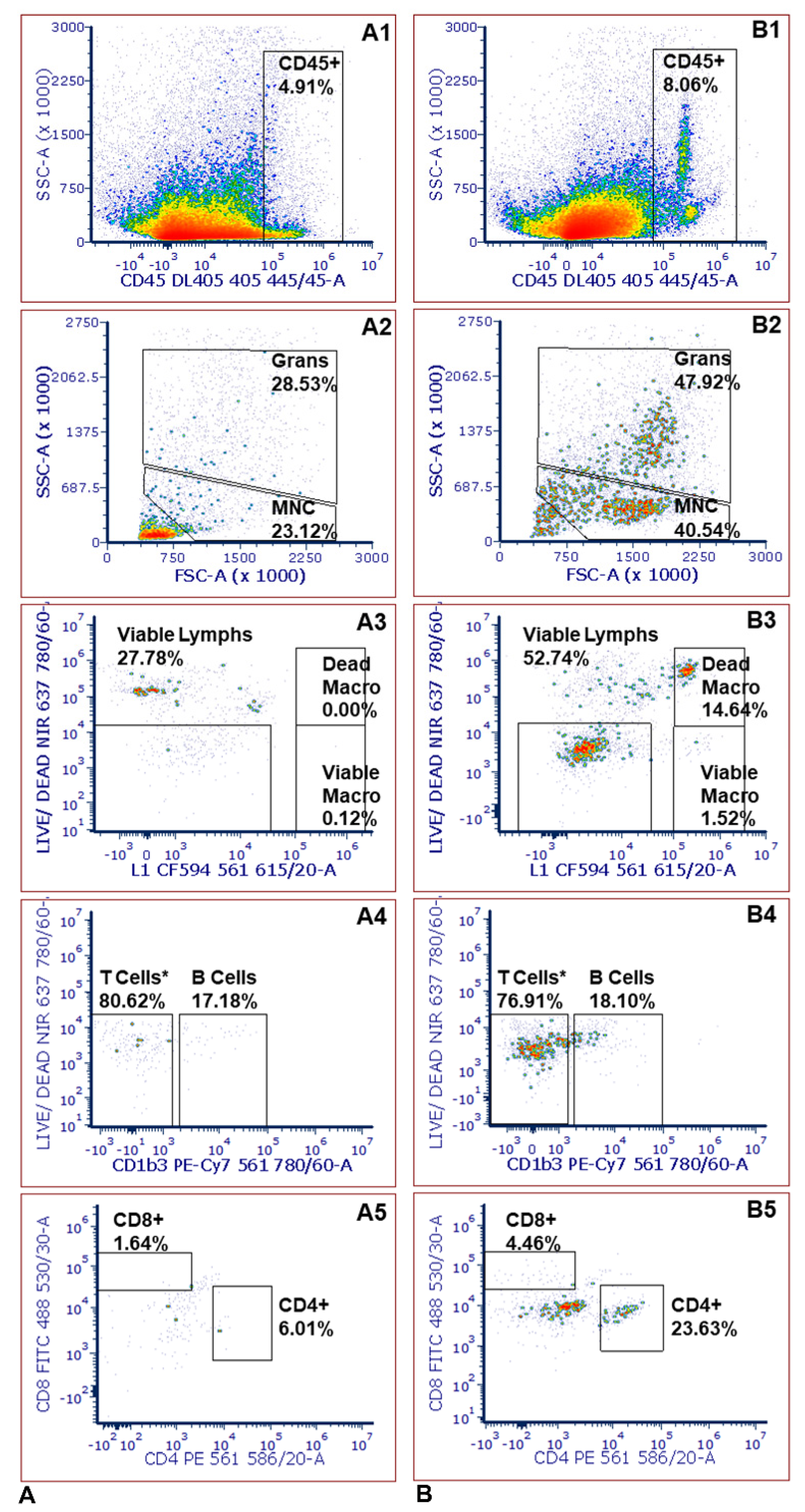

| Replicate Assay 1 | Replicate Assay 2 | Replicate Assay 3 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | Mean | CV | 1 | 2 | 3 | Mean | CV | 1 | 2 | 3 | Mean | CV | |
| B% | 13.8 | 10.7 | 11.5 | 12.0 | 11.1 | 14.4 | 14.4 | 12.9 | 13.9 | 5.0 | 17.1 | 17.1 | 18.0 | 17.4 | 2.3 |
| CD4% | 39.5 | 32.3 | 36.2 | 36.0 | 8.2 | 38.3 | 36.8 | 35.8 | 37.0 | 2.8 | 32.7 | 32.6 | 32.2 | 32.5 | 0.6 |
| CD8% | 15.8 | 16.7 | 17.2 | 16.5 | 3.5 | 28.8 | 29.7 | 29.9 | 29.4 | 1.6 | 19.0 | 18.4 | 18.2 | 18.5 | 1.9 |
| CD4/CD8 | 2.5 | 1.9 | 2.1 | 2.2 | 10.9 | 1.3 | 1.2 | 1.2 | 1.3 | 4.4 | 1.7 | 1.8 | 1.8 | 1.8 | 1.6 |
| Monocyte% | 39.0 | 43.5 | 44.0 | 42.2 | 5.3 | 33.2 | 32.0 | 29.5 | 31.6 | 4.9 | 45.9 | 47.3 | 47.3 | 46.8 | 1.4 |
| Granulocyte% | 0.81 | 0.68 | 0.88 | 0.79 | 10.5 | 0.53 | 0.72 | 0.73 | 0.66 | 13.9 | 0.94 | 0.77 | 1.08 | 0.93 | 13.6 |
| Assay 1 | Assay 2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tick | No Tick | Tick | No Tick | |||||||||||
| 1 | 2 | 3 | Mean | 1 | 2 | Mean | 1 | 2 | 3 | Mean | 1 | 2 | Mean | |
| CD45 (Cell Count) | 10,626 | 5996 | 7976 | 8199 | 3533 | 3439 | 3486 | 3894 | 14,000 | 3847 | 7247 | 1901 | 3061 | 2481 |
| B% | 11.3 | 18.1 | 10.5 | 13.3 | 17.2 | 9.8 | 13.5 | 20.8 | 15.1 | 19.3 | 18.4 | 16.4 | 20.3 | 18.3 |
| CD4% | 28.8 | 23.2 | 34.4 | 28.8 | 6.6 | 4.4 | 5.5 | 19.2 | 28.8 | 13.2 | 20.4 | 3.4 | 4.2 | 3.8 |
| CD8% | 6.6 | 4.5 | 8.9 | 6.7 | 1.64 | 2.9 | 2.3 | 17.3 | 10.1 | 12.2 | 13.2 | 19.7 | 8.4 | 14.0 |
| CD4/CD8 | 4.4 | 5.2 | 3.9 | 4.5 | 4.0 | 1.5 | 2.8 | 1.1 | 2.8 | 1.1 | 1.7 | 0.2 | 0.5 | 0.3 |
| Viable Macrophage% | 0.51 | 1.52 | 0.50 | 0.84 | 0.12 | 0.00 | 0.06 | 0.24 | 1.06 | 1.75 | 1.02 | 0.34 | 0.00 | 0.17 |
| Dead Macrophage% | 17.57 | 14.64 | 1.09 | 11.10 | 0.00 | 0.11 | 0.06 | 1.59 | 46.00 | 1.28 | 16.29 | 2.07 | 0.77 | 1.42 |
| Granulocyte% | 42.4 | 47.9 | 35.4 | 41.9 | 28.53 | 24.1 | 26.3 | 26.5 | 33.5 | 35.3 | 31.8 | 25.5 | 18.7 | 22.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cross, C.E.; Stokes, J.V.; Alugubelly, N.; Ross, A.-M.L.; Willeford, B.V.; Walker, J.D.; Varela-Stokes, A.S. Skin in the Game: An Assay to Monitor Leukocyte Infiltration in Dermal Lesions of a Guinea Pig Model for Tick-Borne Rickettsiosis. Pathogens 2022, 11, 119. https://doi.org/10.3390/pathogens11020119
Cross CE, Stokes JV, Alugubelly N, Ross A-ML, Willeford BV, Walker JD, Varela-Stokes AS. Skin in the Game: An Assay to Monitor Leukocyte Infiltration in Dermal Lesions of a Guinea Pig Model for Tick-Borne Rickettsiosis. Pathogens. 2022; 11(2):119. https://doi.org/10.3390/pathogens11020119
Chicago/Turabian StyleCross, Claire E., John V. Stokes, Navatha Alugubelly, Anne-Marie L. Ross, Bridget V. Willeford, Jamie D. Walker, and Andrea S. Varela-Stokes. 2022. "Skin in the Game: An Assay to Monitor Leukocyte Infiltration in Dermal Lesions of a Guinea Pig Model for Tick-Borne Rickettsiosis" Pathogens 11, no. 2: 119. https://doi.org/10.3390/pathogens11020119
APA StyleCross, C. E., Stokes, J. V., Alugubelly, N., Ross, A.-M. L., Willeford, B. V., Walker, J. D., & Varela-Stokes, A. S. (2022). Skin in the Game: An Assay to Monitor Leukocyte Infiltration in Dermal Lesions of a Guinea Pig Model for Tick-Borne Rickettsiosis. Pathogens, 11(2), 119. https://doi.org/10.3390/pathogens11020119








