Journal Description
Viruses
Viruses
is a peer-reviewed, open access journal of virology, published monthly online by MDPI. The Spanish Society for Virology (SEV), Canadian Society for Virology (CSV), Italian Society for Virology (SIV-ISV), Australasian Virology Society (AVS), Brazilian Society for Virology (BSV) and others are affiliated with Viruses and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Virology) / CiteScore - Q1 (Virology/Infectious Diseases)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 18.6 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Companion journal: Zoonotic Diseases.
Impact Factor:
3.5 (2024);
5-Year Impact Factor:
3.7 (2024)
Latest Articles
Comparative Analysis Reveals Host Species-Dependent Diversity Among 16 Virulent Bacteriophages Isolated Against Soybean Bradyrhizobium spp.
Viruses 2025, 17(11), 1474; https://doi.org/10.3390/v17111474 (registering DOI) - 4 Nov 2025
Abstract
Phages play a role in shaping ecosystems by controlling host abundance via cell lysis, driving host evolution via horizontal gene transfer, and promoting nutrient cycling. The genus Bradyrhizobium includes bacteria able to symbiotically nodulate the roots of soybean (Glycine max), providing
[...] Read more.
Phages play a role in shaping ecosystems by controlling host abundance via cell lysis, driving host evolution via horizontal gene transfer, and promoting nutrient cycling. The genus Bradyrhizobium includes bacteria able to symbiotically nodulate the roots of soybean (Glycine max), providing the plant with a direct source of biologically fixed nitrogen. Optimizing this symbiosis can minimize the use of nitrogen fertilizers and make soybean production more sustainable. Phages targeting Bradyrhizobium may modify their hosts’ genotype, alter phenotypic traits such as symbiotic effectiveness, and mediate competition among strains for nodulation sites. Sixteen phages were isolated against B. diazoefficiens strain USDA110 and B. elkanii strains USDA94 and USDA31. Comparative analyses revealed host species-dependent diversity in morphology, host range, and genome composition, leading to the identification of three previously undescribed phage species. Remarkably, all B. elkanii phages shared a siphophage morphology and formed a single species with >97% nucleotide identity, even when isolated from farms separated by up to ~70 km, suggesting genomic stability across geographic scales. In contrast, phages isolated against B. diazoefficiens had a podophage-like morphology, exhibited greater genetic diversity, and divided into two distinct species. Although no phages were recovered against the B. japonicum strains or native Delaware Bradyrhizobium isolates tested, some Delaware Bradyrhizobium isolates showed susceptibility in a host range assay. The phage genomes demonstrated features predicting phenotypes. The phage terminase genes predicted headful packaging which promotes generalized transduction. The B. elkanii phages all carried tmRNA genes capable of rescuing stalled ribosomes, and all but one of the phages isolated against the two host species carried DNA polymerase A indicating greater phage control of genome replication. State-of-the-art structural annotation of a hypothetical gene shared by the B. diazoefficiens phages, having a mean amino acid identity of ~25% and similarity of ~35%, predicted a putative tail fiber function. Together this work expands the limited knowledge available on soybean Bradyrhizobium phage ecology and genomics.
Full article
(This article belongs to the Section Bacterial Viruses)
Open AccessArticle
Multivalent Interactions Between the Picornavirus 3C(D) Main Protease and RNA Oligonucleotides Induce Liquid–Liquid Phase Separation
by
Somnath Mondal, Saumyak Mukherjee, Kevin E. W. Namitz, Neela H. Yennawar and David D. Boehr
Viruses 2025, 17(11), 1473; https://doi.org/10.3390/v17111473 - 4 Nov 2025
Abstract
The picornavirus 3CD protein is a precursor to the 3C main protease and the 3D RNA-dependent RNA polymerase. In addition to its functions in proteolytic processing of the virus polyprotein and cleavage of key host factors, the 3C domain interacts with cis-acting replication
[...] Read more.
The picornavirus 3CD protein is a precursor to the 3C main protease and the 3D RNA-dependent RNA polymerase. In addition to its functions in proteolytic processing of the virus polyprotein and cleavage of key host factors, the 3C domain interacts with cis-acting replication elements (CREs) within the viral genome to regulate replication and translation events. We investigated the molecular determinants of RNA binding to 3C using a wide range of biophysical and computational methods. These studies showed that 3C binds to a broad spectrum of RNA oligonucleotides, displaying minimal sequence and structure dependence, at least for these shorter RNAs. However, they also uncovered a novel aspect of these interactions, that is, 3C-RNA binding can induce liquid–liquid phase separation (LLPS), with 3CD–RNA interactions likewise leading to LLPS. This may be a general phenomenon for other 3C and 3C-like proteases and polyproteins incorporating 3C domains. These findings have potential implications in understanding virally induced apoptosis and the control of stress granules, which involve LLPS and include other proteins with known interactions with 3C/3CD.
Full article
(This article belongs to the Section General Virology)
►▼
Show Figures
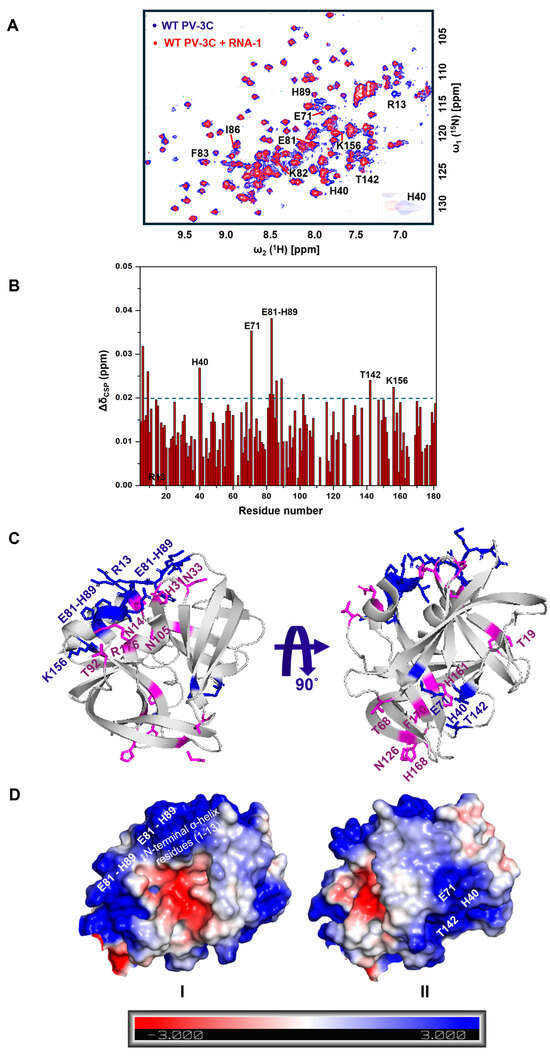
Figure 1
Open AccessArticle
Kinetics of Viral Genome Distribution in Swine Peripheral Lymphoid Organs Following Oronasal Infection with Attenuated African Swine Fever Strains
by
Kalhari Goonewardene, Carissa Embury-Hyatt, Estella Moffat and Aruna Ambagala
Viruses 2025, 17(11), 1472; https://doi.org/10.3390/v17111472 - 4 Nov 2025
Abstract
African swine fever (ASF) continues to spread across the globe, causing a severe impact on the swine industry. Passive surveillance based on testing dead pigs is one of the most effective methods for early detection of ASF incursions. We have previously shown that
[...] Read more.
African swine fever (ASF) continues to spread across the globe, causing a severe impact on the swine industry. Passive surveillance based on testing dead pigs is one of the most effective methods for early detection of ASF incursions. We have previously shown that the superficial inguinal lymph node (SILN) is a convenient and effective sample type for ASF virus (ASFV) genome detection in pigs succumbed to highly or moderately virulent ASFV infections. In this study, we explored the distribution kinetics of ASFV into SILN and other lymphoid tissues in pigs exposed to moderately virulent ASFV strains (ASFV Estonia 2014 and ASFV Malta’78), oronasally. The ASFV genome was detected in SILNs as early as 2–3 days post-infection (dpi), peaking around 5–9 dpi. The detection of ASFV Estonia 2014 started early, and the pigs succumbed to infection faster compared to the ASFV Malta’78 infected pigs that remained longer. All pigs that succumbed to ASF had comparable levels of ASFV genomic material in the spleen and SILNs. The levels of ASFV genomic material gradually started to decrease in pigs that did not succumb to ASF, indicating possible virus clearance. In contrast, ASFV genome levels in blood and spleen samples remained relatively steady during the study period. Immunohistochemistry and in situ hybridization of spleen and SILN samples supported real-time PCR results. This study demonstrates the distribution kinetics of moderately virulent ASFV in peripheral lymph nodes and highlights the utility of SILNs for dead pig screening.
Full article
(This article belongs to the Special Issue ASFV Countermeasures, Pathogenesis, and Epidemiology)
►▼
Show Figures
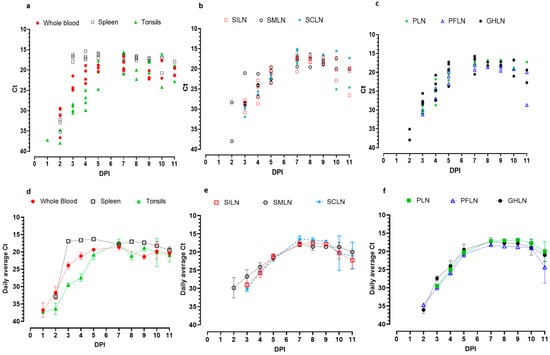
Figure 1
Open AccessArticle
Evaluation of the Efficacy of Tenofovir Alafenamide in Patients with Low-Level Viremia Under Chronic Hepatitis B Treatment
by
Alper Tahmaz, Merve Yıldız Dikmen, Figen Yıldırım, Türkkan Öztürk Kaygusuz, Oğuz Karabay, Mustafa Kemal Çelen, Sevil Alkan, Tuba Damar Çakırca, Fethiye Akgül, Sıla Akhan, Esra Gürbüz, Şafak Özer Balin, Mehmet Çelik and Mehmet Çabalak
Viruses 2025, 17(11), 1471; https://doi.org/10.3390/v17111471 - 4 Nov 2025
Abstract
In this multicenter, retrospective study involving 62 patients, we investigated whether switching from entecavir (ETV) or tenofovir disoproxil fumarate (TDF) to tenofovir alafenamide (TAF) represents a superior treatment strategy for patients with chronic hepatitis B (CHB) experiencing low-level viremia (LLV). The study determined
[...] Read more.
In this multicenter, retrospective study involving 62 patients, we investigated whether switching from entecavir (ETV) or tenofovir disoproxil fumarate (TDF) to tenofovir alafenamide (TAF) represents a superior treatment strategy for patients with chronic hepatitis B (CHB) experiencing low-level viremia (LLV). The study determined that TAF significantly improved both virological and biochemical outcomes. At 48 weeks, the complete virological response (CVR) rate was 77.8% for those who switched from ETV and 81.8% for those who switched from TDF, with Hepatitis B virus deoxyribonucleic acid (HBV DNA) negativity reaching 81% by month 12. Additionally, significant normalization of liver enzymes, albumin, and platelet counts was observed across the cohort. While the switch from TDF was associated with a significant increase in triglycerides and high-density lipoprotein (HDL) and a decrease in estimated glomerular filtration rate (eGFR), no such changes were detected in the ETV group. This evidence suggests that TAF provides robust virological control in LLV patients and is associated with favorable biochemical improvements. However, due to the study’s limitations, the strong assertion that TAF promotes the regression of liver fibrosis and reduces the risk of hepatocellular carcinoma (HCC) must be interpreted with caution.
Full article
(This article belongs to the Section Viral Immunology, Vaccines, and Antivirals)
Open AccessArticle
Comparative Analysis of Extracellular Vesicle and Virus Co-Purified Fractions Produced by Contemporary Influenza A and B Viruses in Different Human Cell Lines
by
Aude Wantchecon, Julien Boucher, Henintsoa Rabezanahary, Caroline Gilbert and Mariana Baz
Viruses 2025, 17(11), 1470; https://doi.org/10.3390/v17111470 - 4 Nov 2025
Abstract
Influenza virus is one of the most frequent causes of respiratory infection in humans. Recent studies suggest that extracellular vesicles (EVs)—small particles released by cells during influenza virus infection—can influence the immune response and viral pathogenesis. However, during viral replication, infected cells can
[...] Read more.
Influenza virus is one of the most frequent causes of respiratory infection in humans. Recent studies suggest that extracellular vesicles (EVs)—small particles released by cells during influenza virus infection—can influence the immune response and viral pathogenesis. However, during viral replication, infected cells can also release EVs, which may include different subtypes. This study aimed to purify and characterize viral preparations and EVs using sequential ultracentrifugation methods. Influenza A/H1N1, A/H3N2, and B virus strains were produced in human Calu-3 and A549 cell lines. Viral supernatants then underwent a series of differential ultracentrifugation steps at 3000× g, 17,000× g, and 100,000× g. Dynamic light scattering analysis (DLS) validated size heterogeneity for all three types of EVs. Measurement of infectious virus particles for all three pellets showed virus enrichment at 17,000× g and 100,000× g. Dot blot analysis confirmed the enrichment of virus particles in these fractions and the presence of EV protein. This study demonstrates the presence of EVs in virus preparations and highlights the need for improved separation methods to characterize them better and explore their role in viral infection pathogenesis.
Full article
(This article belongs to the Special Issue Women in Virology 2025)
►▼
Show Figures
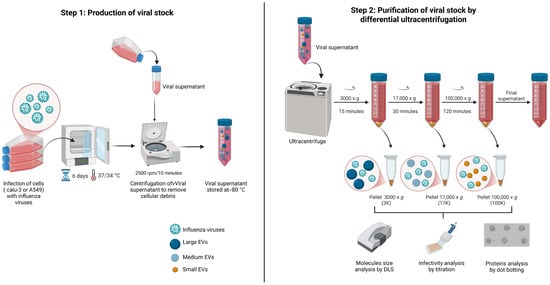
Figure 1
Open AccessReview
The Role of Innate Cells During Alphavirus Chikungunya Infection
by
Juliane Santos de França da Silva, Livian Maria Silva dos Santos, Célio Valdevino Ferreira Junior, Nathalie de Sena Pereira, Juliana Navarro Ueda Yaochite, Valter Ferreira de Andrade Neto, Paulo Marcos da Matta Guedes, Rafael Freitas De Oliveira França, Ramayana Morais de Medeiros Brito and Manuela Sales Lima Nascimento
Viruses 2025, 17(11), 1469; https://doi.org/10.3390/v17111469 - 1 Nov 2025
Abstract
Alphavirus chikungunya (CHIKV) is an arthropod-borne alphavirus of the Togaviridae family, transmitted primarily by Aedes aegypti and Ae. albopictus mosquitoes. CHIKV infection often results in debilitating manifestations that compromise quality of life and generate significant socioeconomic impacts. Recurrent epidemics in tropical and subtropical
[...] Read more.
Alphavirus chikungunya (CHIKV) is an arthropod-borne alphavirus of the Togaviridae family, transmitted primarily by Aedes aegypti and Ae. albopictus mosquitoes. CHIKV infection often results in debilitating manifestations that compromise quality of life and generate significant socioeconomic impacts. Recurrent epidemics in tropical and subtropical regions underscore the urgent need to better understand the host immune responses and their contribution to disease outcome. CHIKV establishes infection by overcoming the host’s initial immunological barriers. Innate immune cells, including fibroblasts, dendritic cells, macrophages, monocytes, neutrophils and natural killer (NK) cells, are among the first to respond to infection, ensuring a rapid antiviral defense and supporting the development of adaptive immune responses. However, excessive release of inflammatory mediators and prolonged infiltration of innate cells into joint tissues contribute to disease chronicity and the persistence of arthralgia. In this review, we provide a comprehensive synthesis of current evidence on innate cells that serve as targets for CHIKV infection, highlighting mechanisms that promote effective antiviral defense as well as those responsible for pathological inflammation and chronic disease and identifying key gaps that remain to be addressed.
Full article
(This article belongs to the Special Issue Advances in Understanding Viral Pathogenesis and Host Immune Responses to Arboviruses and Respiratory Viruses)
►▼
Show Figures
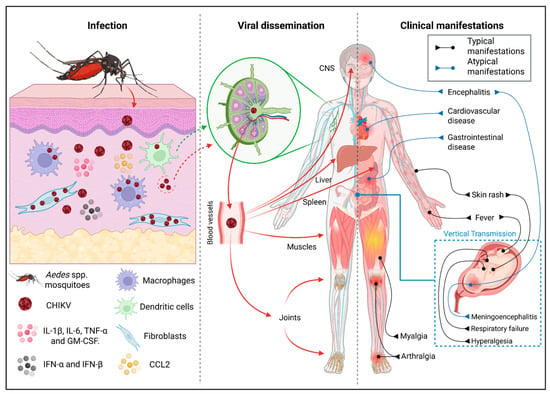
Figure 1
Open AccessArticle
ORF3 Gene of Porcine Epidemic Diarrhea Virus Causes Nuclear and Morphological Distortions with Associated Cell Death
by
Ndirangu A. Kamau, Jae-Rang Rho, Eui-Soon Park, Jung-Eun Yu, Ji-Yun Yu, Gianmarco Ferrara and Hyun-Jin Shin
Viruses 2025, 17(11), 1468; https://doi.org/10.3390/v17111468 - 1 Nov 2025
Abstract
There is increasing research interest in the ORF3 accessory protein of PEDV as a critical element for viral virulence. Here, wild type ORF3 (ORF3wt) gene was constructed in pEGFP-C1 vector. Additionally, two truncation mutants, ORF3-N (1-98 amino acids [aa]) and ORF3-C
[...] Read more.
There is increasing research interest in the ORF3 accessory protein of PEDV as a critical element for viral virulence. Here, wild type ORF3 (ORF3wt) gene was constructed in pEGFP-C1 vector. Additionally, two truncation mutants, ORF3-N (1-98 amino acids [aa]) and ORF3-C (99-224 aa) were inserted in the same vector. Results of ORF3 expression revealed early cytoplasmic localization but 12 h after transfection, ORF3 accumulated around the nucleus, especially ORF3-N. This caused chromosome condensation and morphological distortion that culminated in cell death. In comparison with the native cells expressing GFP alone, ORF3wt-induced lethality was 6.61% above baseline while ORF3- C expression resulted in moderate increase in cell death (0.64%). ORF3-N was affected the most with 220.32% increased lethality. It was, therefore, inferred that the ORF3 gene encodes a protein that causes nuclear damage, distorts cell morphology and leads to cell death. Furthermore, the role of the protein could be inherent in the N-terminal domain, which consists of the transmembrane domains. These findings underpin the importance of ORF3 gene expression in the host and are rudimental insights for further exploration into the mechanistic interactions of ORF3 and the host, as well as a possible role in pathogenesis in PEDV and other coronaviruses.
Full article
(This article belongs to the Special Issue Porcine Epidemic Diarrhea Virus (PEDV): A Persistent Threat to the Global Swine Industry)
►▼
Show Figures

Figure 1
Open AccessArticle
Clinical Characteristics and Outcomes of ICU Patients During the First Post-COVID-19 2023–2024 Influenza Season in The Netherlands
by
Sjoerd van der Bie, Johannes P. C. van den Akker, Ramon C. Fluit, Steven F. L. van Lelyveld, Maarten E. Nuver, Suzanne Stads, Peter Spronk, Carina Bethlehem, Romy Takken, Corstiaan A. den Uil, Jantine Van Holten, Rutger Van Raalte, Jurre Kuipers, Marc Schluep, Matty Koopmans, Louise Urlings-Strop, Esther K. Haspels-Hogervorst, Nina E. Disseldorp, Jan Elderman, Roy Sneijder, Jasper de Roos, Merijn Kant, Robbert G. Bentvelsen, Tobias Neijzen, Dorien Kiers, Klaas de Groot, Ashley de Bie, Peter de Jager, Michiel Blans, Myrthe de Haas, Mariska Lont, Stephanie Koster, Angelique C. M. Jansen, Petronella E. Deetman, Fieke Mus, Ralph Nowitzky, Lucas Brands, Hazra Moeniralam, Erik Schaftenaar, Martijn van Tellingen, Jasper Haringman, Emily Thieme Groen, Lenneke E. M. Haas, Wouter de Ruijter, Rob Wilting, Hetty Kranen, Charlotte H. S. B. van den Berg, Diederik Gommers, Evert-jan Wils, Henrik Endeman and Marco Goeijenbieradd
Show full author list
remove
Hide full author list
Viruses 2025, 17(11), 1467; https://doi.org/10.3390/v17111467 - 1 Nov 2025
Abstract
Background: Influenza can cause severe complications, especially in patients with specific risk factors or comorbidities associated with poor outcomes. Some patients are at increased risk of a complicated disease course, including secondary infections, ICU admission, and the need for mechanical ventilation. The first
[...] Read more.
Background: Influenza can cause severe complications, especially in patients with specific risk factors or comorbidities associated with poor outcomes. Some patients are at increased risk of a complicated disease course, including secondary infections, ICU admission, and the need for mechanical ventilation. The first post–COVID-19 seasonal influenza season placed a substantial burden on Dutch ICUs. This study investigates the disease course and outcomes of ICU patients with influenza. Methods: A retrospective influenza registry study was conducted across 34 Dutch ICUs, including patients aged 18 and older admitted to the ICU with a positive influenza RT-PCR test, between 1 November 2023 and 17 March 2024. Data on demographic information, medical history, clinical symptoms, laboratory and imaging results, parameters of mechanical ventilation, additional treatments, length of hospital stay, and mortality was retrieved from the electronic patient record. Results: A total of 498 patients were included in the study. The median age was 64 (IQR: 55–72) years and 58.8% of the patients were men. The most common comorbidities were cardiovascular disease (34.1%), chronic obstructive pulmonary disease (COPD) (31.5%), and diabetes (22.3%). Bacterial co-infections were present in 37.6% of the patients. Invasive mechanical ventilation (IMV) was necessary in 46.0% of patients, 38.0% of those requiring IMV were treated in prone position. A substantial mortality rate was observed, with an ICU mortality rate of 21.9% and an additional hospital mortality rate of 5.2%. Conclusion: This study described the characteristics and course of disease of all patients with laboratory-confirmed influenza infection admitted to one of the 34 participating Dutch ICUs between November 2023 and March 2024. The major findings of this study are the substantial mortality rate, a high proportion of patients with bacterial co-infections, and a significant percentage of patients requiring IMV and prone position ventilation. Finally, patients without comorbidities that were admitted to the ICU with an influenza virus infection showed severe disease parameters but had a lower mortality than patients with comorbidities.
Full article
(This article belongs to the Section Human Virology and Viral Diseases)
Open AccessArticle
Lake Trout (Salvelinus namaycush) Naturally Infected with Salmovirus salmonidallo3 (SalHV-3; Family Alloherpesviridae) Continue to Harbor the Virus for Nearly a Decade
by
Megan A. Shavalier, Mohamed Faisal and Thomas P. Loch
Viruses 2025, 17(11), 1466; https://doi.org/10.3390/v17111466 - 31 Oct 2025
Abstract
Salmovirus salmonidallo3 (SalHV-3) causes Epizootic Epitheliotropic Disease (EED), which has resulted in the death of millions of lake trout (Salvelinus namaycush) over the past 40 years. Although advancements in understanding this virus’s pathogenicity and control strategies have been made, the duration
[...] Read more.
Salmovirus salmonidallo3 (SalHV-3) causes Epizootic Epitheliotropic Disease (EED), which has resulted in the death of millions of lake trout (Salvelinus namaycush) over the past 40 years. Although advancements in understanding this virus’s pathogenicity and control strategies have been made, the duration and effects of chronic SalHV-3 infections remain unknown. This study focused on lake trout that survived a natural outbreak of EED in 2012 and were maintained under quarantine conditions until 2021. Following exposure to either repeated or intermittent handling stress designed to mimic typical hatchery practices, SalHV-3 was detected (via a SalHV-3-specific quantitative PCR assay) in multiple tissues from multiple fish. Non-lethally collected tissues revealed the highest prevalence and virus loads in the fin and mucus. SalHV-3 was detected in these tissues throughout the study period (49 days, 8 sampling events), with some fish having detectible virus on each study day and others only intermittently (n = 1–7 sampling days). Tissues collected lethally yielded SalHV-3 detections in multiple nervous tissues, as well as in the cornea of several fish. Experiments to evaluate virus shedding revealed that SalHV-3 was intermittently detectable in fish holding water. Collectively, results indicate that lake trout can remain SalHV-3 infected for nearly a decade and intermittently shed the virus, constituting a threat to hatchery-based lake trout conservation efforts in the Great Lakes basin.
Full article
(This article belongs to the Special Issue Aquatic Animal Viruses and Antiviral Immunity)
►▼
Show Figures
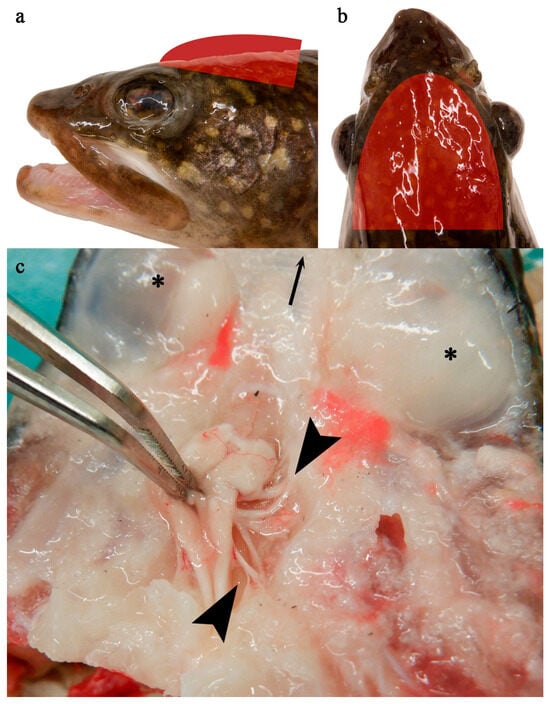
Figure 1
Open AccessArticle
Evaluation of a Probe-Based Enrichment Protocol for Nanopore Sequencing of Zoonotic Viruses
by
Kailin Hawes, Benjamin Greene, Zachary A. Weishampel, Paul A. Beare, Sarah van Tol, Paul Schaughency, Skyler Kuhn, Alison J. Peel, Vincent J. Munster and Claude Kwe Yinda
Viruses 2025, 17(11), 1465; https://doi.org/10.3390/v17111465 - 31 Oct 2025
Abstract
The detection of high-consequence viral pathogens is essential for spillover prevention and reduction in transmission but is limited by the low sensitivity of next-generation sequencing technology. Low-titer field samples from a variety of hosts are primarily composed of non-viral genomic material, reducing the
[...] Read more.
The detection of high-consequence viral pathogens is essential for spillover prevention and reduction in transmission but is limited by the low sensitivity of next-generation sequencing technology. Low-titer field samples from a variety of hosts are primarily composed of non-viral genomic material, reducing the probability of obtaining usable sequence data. Targeted enrichment, such as VirCapSeq-VERT, removes background genomic material to improve virus detection but is mainly used for sequencing clinical samples. We customized the VirCapSeq-VERT probe system to aid in the detection of zoonotic viruses of interest and adapted it for use on the Oxford Nanopore sequencing platform. We validated the method on a variety of samples, including a mock virome consisting of seven RNA viruses, samples from an animal laboratory study, and a set of animal field samples. We also developed Nanite, a lightweight bioinformatics pipeline, to perform bioinformatic analyses. Results indicated that the optimized enrichment protocol improved sequencing by enhancing the detection of viruses, increasing read lengths, and, in some cases, improving genomic coverage. Most importantly, the sequencing of zoonotic viruses was improved in field samples with low titers, suggesting that this protocol is a useful tool for increasing the efficacy of Oxford Nanopore sequencing for field-oriented applications.
Full article
(This article belongs to the Section General Virology)
►▼
Show Figures
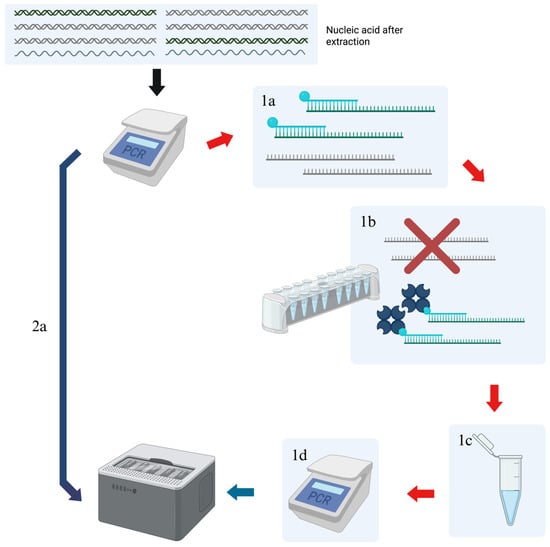
Figure 1
Open AccessArticle
Antiviral Activity of Essential Oils Against Avian Influenza Virus H7N3 In Vitro and In Ovo Models
by
Inkar Castellanos-Huerta, Jaime A. Ángel-Isaza, Lucio Bacab-Cab, Kevin Yam-Trujillo, Alejandro Aranda, Sindi Alejandra Velandia-Cruz, Loufrantz Mendez, Victor M. Petrone-Garcia, Guadalupe Ayora-Talavera and Álvaro José Uribe
Viruses 2025, 17(11), 1464; https://doi.org/10.3390/v17111464 - 31 Oct 2025
Abstract
The poultry industry is continually seeking efficient, practical strategies to control infectious diseases. Among these new alternatives are essential oils (EOs), naturally occurring compounds with antimicrobial properties. Their effectiveness has been demonstrated in various studies that focus on their broad antiviral properties. The
[...] Read more.
The poultry industry is continually seeking efficient, practical strategies to control infectious diseases. Among these new alternatives are essential oils (EOs), naturally occurring compounds with antimicrobial properties. Their effectiveness has been demonstrated in various studies that focus on their broad antiviral properties. The present experiment evaluated the antiviral efficacy of an EOs formulation against the H7N3 subtype of avian influenza virus (AIV) by directly mixing virus and EOs (virus/EOs mixture) through an in vitro model in cultured Madin-Darby canine kidney cells (MDCKs). The experiment used a focus reduction neutralization test (FRNT) to determine the 50% inhibitory concentration (IC50) by virus/EOs mixture, as well as the application of EOs 24 h before infection, through a viral inhibition test using a chicken embryo (CE) in ovo model. The results demonstrated the antiviral activity of the EOs formulation against the H7N3 in vitro model (IC50 values of 20.4 and 38.3 ppm and selective index (SI) values of 9.4 and 5.1) and in ovo model (decreasing hemagglutination titers to 1 HA unit, 105.28 embryo infectious dose 50% (EID50) per mL, and viral loads to approximately 1011.4 copies/mL) when applied in CE, 24 h before viral infection, representing the lowest replication indicators recorded during the experiment. According to the results, the EOs formulation demonstrated antiviral activity against AIV H7N3 both as a virus/EOs mixture and through application in ovo 24 h before infection. Application 24 h before infection in CE showed a significant effect compared with the virus/EOs mixture, demonstrating an antiviral effect in the ovo infection model. This study demonstrates both the virucidal and antiviral capacity of the compounds in the EOs formulation against AIV H7N3 and their efficacy when applied 24 h before infection in the in ovo model.
Full article
(This article belongs to the Special Issue Antiviral Agents to Influenza Virus 2025)
►▼
Show Figures

Graphical abstract
Open AccessReview
Combating White Spot Syndrome Virus (WSSV) in Global Shrimp Farming: Unraveling Its Biology, Pathology, and Control Strategies
by
Md. Iftehimul, Neaz A. Hasan, David Bass, Abul Bashar, Mohammad Mahfujul Haque and Morena Santi
Viruses 2025, 17(11), 1463; https://doi.org/10.3390/v17111463 - 31 Oct 2025
Abstract
White Spot Syndrome Virus (WSSV) is one of the most devastating viral pathogens affecting shrimp, causing severe economic losses to the global farmed shrimp trade. The globalization of live shrimp trade and waterborne transmission have facilitated the rapid spread of WSSV across major
[...] Read more.
White Spot Syndrome Virus (WSSV) is one of the most devastating viral pathogens affecting shrimp, causing severe economic losses to the global farmed shrimp trade. The globalization of live shrimp trade and waterborne transmission have facilitated the rapid spread of WSSV across major shrimp-producing countries since its initial emergence. The present review gives an updated account of WSSV biology, pathology, transmission dynamics, and recent developments in control measures. The virus, a double-stranded DNA virus of the Nimaviridae family, utilizes advanced immune evasion strategies, resulting in severe mortality. Shrimp lack adaptive immunity and hence rely predominantly on innate immunity, which is insufficient to mount an effective response against severe infections. Traditional disease control measures such as augmented biosecurity, selective breeding, and immunostimulants have, despite extensive research, achieved only limited success. New biotechnological tools such as RNA interference, CRISPR-Cas gene editing, and nanotechnology offer tremendous potential for disease mitigation. In parallel, the development of DNA and RNA vaccines targeting WSSV structural proteins, such as VP28, holds significant promise for stimulating the shrimp immune system. This review highlights the urgent need for a convergent approach to sustainable disease management in global shrimp aquaculture, with interdisciplinarity playing a pivotal role in shaping the future of WSSV control.
Full article
(This article belongs to the Section Viral Immunology, Vaccines, and Antivirals)
►▼
Show Figures
Figure 1
Open AccessCase Report
Reversion of Val(Ganciclovir)-Resistance-Associated Mutations in Two SOT Patients with Mismatched Serostatus for CMV (D+/R-)
by
Elena Seminari, Alessandra Tebaldi, Aurelia Sangani, Paola Giordani, Daniele Lilleri, Stefania Paolucci, Giulia Campanini, Elizabeth Iskandar, Fausto Baldanti and Raffaele Bruno
Viruses 2025, 17(11), 1462; https://doi.org/10.3390/v17111462 - 31 Oct 2025
Abstract
The emergence of drug-resistant cytomegalovirus (CMV) complicates viral response to therapy. We present two cases of solid organ transplant (SOT) recipients, highlighting the reversion of UL97 mutations associated with val(ganciclovir) resistance. Patient 1, a heart transplant recipient, initially received pre-emptive treatment with val(ganciclovir),
[...] Read more.
The emergence of drug-resistant cytomegalovirus (CMV) complicates viral response to therapy. We present two cases of solid organ transplant (SOT) recipients, highlighting the reversion of UL97 mutations associated with val(ganciclovir) resistance. Patient 1, a heart transplant recipient, initially received pre-emptive treatment with val(ganciclovir), followed by foscarnet for recurrent CMV episodes. Mutations A594V in the UL97-kinase gene and V715M in the UL54-polymerase gene were detected. He developed CMV colitis and was then treated with maribavir. After discontinuing val(ganciclovir), genotyping revealed no resistance mutations. Following CMV DNA suppression, secondary prophylaxis with letermovir and val(ganciclovir) was initiated. Patient 2, a double-lung transplant recipient, experienced several CMV episodes. Initially treated with val(ganciclovir), he developed the L595S mutation in the UL97 kinase gene, conferring resistance. Therapy was then switched to foscarnet, which was suspended due to renal failure, and then to maribavir. Subsequently, the H411Y mutation in the UL97 was detected, conferring maribavir resistance, while val(ganciclovir) mutation was no longer detectable. He was then treated with val(ganciclovir) and letermovir, achieving undetectable CMV DNA, and then continued letermovir alone as prophylaxis. Detecting gene mutations that confer drug resistance is crucial for managing antiviral therapy when virological response is lacking. In our cases, the reversion of (va)ganciclovir-resistance mutations occurred after drug withdrawal, a previously unreported finding.
Full article
(This article belongs to the Special Issue Human Cytomegalovirus Therapeutic Strategies and Clinical Applications)
Open AccessArticle
A Multi-Host Approach to Quantitatively Assess the Role of Dogs as Sentinels for Rift Valley Fever Virus (RVFV) Surveillance in Madagascar
by
Herilantonirina Solotiana Ramaroson, Andres Garchitorena, Vincent Lacoste, Soa Fy Andriamandimby, Matthieu Schoenhals, Jonathan Bastard, Katerina Albrechtova, Laure J. G. Chevalier, Domoina Rakotomanana, Patrick de Valois Rasamoel, Modestine Raliniaina, Heritiana Fanomezantsoa Andriamahefa, Mamitiana Aimé Andriamananjara, Lova Tsikiniaina Rasoloharimanana, Solohery Lalaina Razafimahatratra, Claude Arsène Ratsimbasoa, Benoit Durand and Véronique Chevalier
Viruses 2025, 17(11), 1461; https://doi.org/10.3390/v17111461 - 31 Oct 2025
Abstract
Sentinel animals may play a key role in the surveillance of arbovirus circulation, particularly in developing countries. This study aimed to assess the relevance of using dogs as sentinel animals for Rift Valley fever virus (RVFV) surveillance in Madagascar. Serological surveys were conducted
[...] Read more.
Sentinel animals may play a key role in the surveillance of arbovirus circulation, particularly in developing countries. This study aimed to assess the relevance of using dogs as sentinel animals for Rift Valley fever virus (RVFV) surveillance in Madagascar. Serological surveys were conducted on 513 dogs and 135 cattle in the Ifanadiana district, southeastern Madagascar. In addition, 486 human dry blood samples available from the same area were used. Antibodies against RVFV were detected in 23 of 513 dogs, in 86 of 486 humans, and in 33 of 135 cattle. Serocatalytic models fitted to age-stratified serological data were developed to estimate the RVFV force of infection (FOI) under several hypotheses, ranging from no relationship to proportional RVFV FOIs between humans, cattle, and dogs. The best supported model indicated that RVFV FOI in humans and cattle was proportional to RVFV FOI in dogs. Proportionality parameters were estimated at 2.6 (95% credible interval: [1.4–5.1]) for humans and 3.5 (95% credible interval: [1.3–6.4]) for cattle. Our findings suggested that dog blood samples could be used to identify RVFV circulation in RVF endemic areas and infer the exposure of humans and cattle in these areas in Madagascar.
Full article
(This article belongs to the Special Issue Zoonotic and Vector-Borne Viral Diseases)
►▼
Show Figures
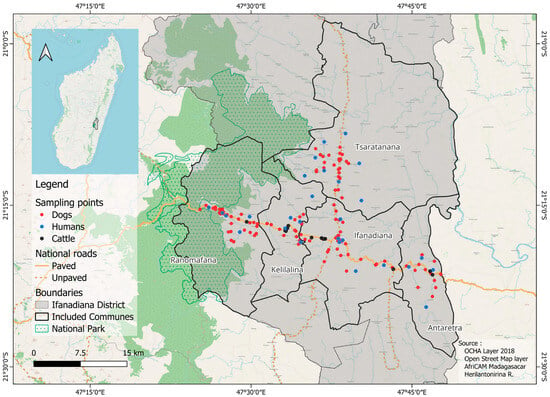
Figure 1
Open AccessArticle
Transcriptomics Reveals the Inhibitory Effect of Scutellarin on PRRSV-Infected PAMs
by
Guidong Zhang, Teng Tu, Yanwei Li, Yueyan Zeng, Mingpeng Hu, Chengchao Du, Zexiao Yang, Xueping Yao, Dishi Chen, Tian Shi and Yin Wang
Viruses 2025, 17(11), 1460; https://doi.org/10.3390/v17111460 - 31 Oct 2025
Abstract
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious epidemic caused by the porcine reproductive and respiratory syndrome virus (PRRSV). Characterized by reproductive disorders in pregnant sows and respiratory symptoms in pigs of all ages, it poses a severe threat to the
[...] Read more.
Porcine reproductive and respiratory syndrome (PRRS) is a highly contagious epidemic caused by the porcine reproductive and respiratory syndrome virus (PRRSV). Characterized by reproductive disorders in pregnant sows and respiratory symptoms in pigs of all ages, it poses a severe threat to the global swine industry. In recent years, the high mutation rate of PRRSV has increasingly limited the effectiveness of vaccines against it, prompting the search for new anti-PRRSV drugs. scutellarin (SCU), a natural flavonoid compound extracted from the medicinal plant Scutellaria baicalensis, possesses multiple biological activities. Its antiviral effects have been demonstrated in numerous studies; however, its inhibitory activity against PRRSV and the underlying mechanism remain unknown. In this study, through in vitro cell experiments, we found that scutellarin significantly inhibits PRRSV infection in PAMs. Furthermore, it directly acts on PRRSV to exert antiviral effects. Transcriptomic analysis suggests that scutellarin may exert its anti-PRRSV effects by regulating host immunity and anti-inflammation through immune-related signaling pathways, including the complement and coagulation cascades, ECM-receptor interaction, Rap1 signaling pathway, and PI3K-Akt signaling pathway.
Full article
(This article belongs to the Section Animal Viruses)
►▼
Show Figures
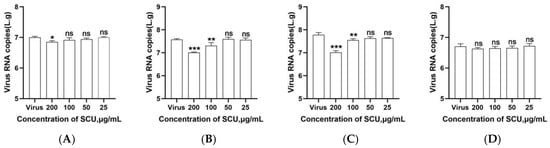
Figure 1
Open AccessArticle
SARS-CoV-2 Serological Surveillance of Both Vaccinated and Unvaccinated Zoo Animals with the Identification of a Sloth Bear and a Tapir with Previous Infection
by
Marie Arvidson, Yashaswi Raj Subedi, Sandipty Kayastha, Angel Mitchell, Kami Alvarado, Xufang Deng, Karen Terio, Matthew Allender and Leyi Wang
Viruses 2025, 17(11), 1459; https://doi.org/10.3390/v17111459 - 31 Oct 2025
Abstract
Since its discovery in 2019, SARS-CoV-2 has continued to be detected in both humans and animals worldwide. Currently there is limited research focusing on serological surveillance of wildlife under human care. Here we tested 230 serum samples of 134 animals from two zoological
[...] Read more.
Since its discovery in 2019, SARS-CoV-2 has continued to be detected in both humans and animals worldwide. Currently there is limited research focusing on serological surveillance of wildlife under human care. Here we tested 230 serum samples of 134 animals from two zoological institutions collected between 2015 and 2024. To assess prior exposure and antibody responses from natural infection or vaccination, we used three serological assays: a nucleocapsid protein-based ELISA (N-ELISA), a surrogate virus neutralization test (sVNT) for spike (S) protein and a neutralization assay with S-pseudotyped viral particles. Among the 114 samples collected from 58 animals at Fort Wayne Zoo in Indiana, 37 samples from 20 vaccinated animals were sVNT-positive, and 2 of the positive animals had 2 samples prior to vaccination that tested positive by N-ELISA. Of the 116 samples from 76 animals at Brookfield Zoo in Illinois, 20 samples of 20 animals were sVNT-positive, and 19 of the positive animals had been vaccinated. Among these 20 sVNT-positive samples, only one sample from a South American Tapir was positive from prior to vaccination and 1 sample from a sloth bear was also positive by N-ELISA, marking the first documented cases of SARS-CoV-2 exposure in both species. Neutralization assays with S-pseudotyped virus revealed that some of the sVNT-positive samples have strong activity against the WH1-S pseudovirus but showed significantly reduced neutralization against the Omicron LP.8.1-S pseudovirus. These results underscore the need for updated vaccines tailored to emerging variants. Overall, our findings highlight the importance of continued serological surveillance across multiple species to detect new SARS-CoV-2 exposures and monitor vaccine-induced immunity in captive animal populations.
Full article
(This article belongs to the Section Coronaviruses)
►▼
Show Figures
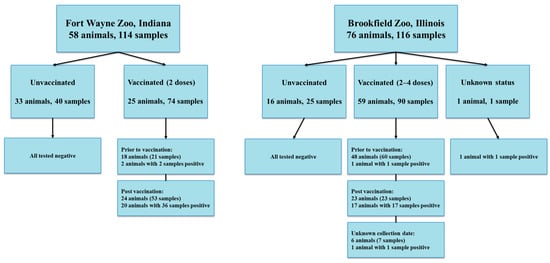
Figure 1
Open AccessArticle
Baculovirus-Mediated Gene Therapy: Targeting BIRC6 for Lung and Breast Cancer
by
Abril Marchesini, Santiago M. Gómez Bergna, Leslie C. Amorós Morales, María Florencia López, Larisa Vásquez, Silvana E. Tongiani, Florencia González Morán, Víctor Romanowski, María Florencia Gottardo and Matias L. Pidre
Viruses 2025, 17(11), 1458; https://doi.org/10.3390/v17111458 - 31 Oct 2025
Abstract
BIRC6, a member of the inhibitor of apoptosis protein family (IAP), regulates apoptosis, autophagy and cytokinesis. IAPs are often overexpressed in tumors, contributing to oncogenesis, therapy resistance and worse prognosis. In particular, BIRC6 overexpression has been found in several tumor tissues. The aim
[...] Read more.
BIRC6, a member of the inhibitor of apoptosis protein family (IAP), regulates apoptosis, autophagy and cytokinesis. IAPs are often overexpressed in tumors, contributing to oncogenesis, therapy resistance and worse prognosis. In particular, BIRC6 overexpression has been found in several tumor tissues. The aim of this study was to evaluate the effect of BIRC6 silencing on the apoptotic response of breast and lung tumor cells. We used RNA interference based on short hairpin RNA (shRNA) to knock down gene expression encoded by a recombinant baculovirus (BV), an insect-specific virus unable to replicate in mammalian hosts, to carry out preclinical validation tests in experimental models both in vitro and in vivo. Our results indicate that BIRC6 plays an antiapoptotic role in both breast and lung tumor cells. In vivo, treatment with BV-shBRIC6 reduced breast and lung tumor progression and increased overall survival. After histological analysis, BV-shBRIC6 was able to increase tumor necrosis. In addition, we demonstrated that BIRC6 expression correlates with antiapoptotic and tumor progression-relevant markers in lung and breast cancer patients. BV-based silencing of BIRC6 may have therapeutic value for the treatment of lung and breast tumors. Further translational studies of BV-shBIRC6 in lung and breast cancer are warranted.
Full article
(This article belongs to the Special Issue Applications of Baculoviruses: Expression Factories, Vaccines and VLPs, Gene Delivery Vectors, and Virus Genetics Models)
►▼
Show Figures

Figure 1
Open AccessReview
Global Surveillance and Biological Characterization of the SARS-CoV-2 NB.1.8.1 Variant: An Emerging VUM Lineage Under Scrutiny
by
Gaojie Cao, Chenhui Xu, Linxi Wang, Keikei Chai and Beibei Wu
Viruses 2025, 17(11), 1457; https://doi.org/10.3390/v17111457 - 31 Oct 2025
Abstract
The continuous evolution of SARS-CoV-2 and its variants poses persistent challenges to global public health. As a sublineage of the XDV.1 variant, NB.1.8.1 has rapidly emerged as a dominant strain worldwide, triggering a new wave of infections. Representing a product of viral adaptation,
[...] Read more.
The continuous evolution of SARS-CoV-2 and its variants poses persistent challenges to global public health. As a sublineage of the XDV.1 variant, NB.1.8.1 has rapidly emerged as a dominant strain worldwide, triggering a new wave of infections. Representing a product of viral adaptation, this variant has acquired several critical amino acid mutations—including A435S and T478I—which enhance its transmissibility and immune evasion capabilities compared to the ancestral XDV.1 lineage. This review systematically summarizes the genomic characteristics, epidemiological features, and immune escape potential of NB.1.8.1. It emphasizes that sustained genomic surveillance and serological assessments are crucial for informing public health response strategies, guiding vaccine development, and optimizing containment measures.
Full article
(This article belongs to the Special Issue Molecular Epidemiology of SARS-CoV-2, 4th Edition)
►▼
Show Figures

Figure 1
Open AccessArticle
Functional Analysis of the Pathogenesis-Related Protein 1 (CaPR1) Gene in the Pepper Response to Chilli veinal mottle virus (ChiVMV) Infection
by
Chunzi Huang, Zengjing Zhao, Xing Wu, Hu Zhao, Meng Wang, Zhi He, Zongjun Li, Lihao Wang, Yafei Tang, Risheng Wang, Longfei He and Mingxia Gong
Viruses 2025, 17(11), 1456; https://doi.org/10.3390/v17111456 - 31 Oct 2025
Abstract
Chilli veinal mottle virus (ChiVMV) causes severe yield losses in pepper across Asia. It is very urgent to study the host plant resistance to control this viral disease. As a type of defense response gene, pathogenesis-related protein 1 (PR1) is a well-established defense
[...] Read more.
Chilli veinal mottle virus (ChiVMV) causes severe yield losses in pepper across Asia. It is very urgent to study the host plant resistance to control this viral disease. As a type of defense response gene, pathogenesis-related protein 1 (PR1) is a well-established defense marker against fungal/bacterial pathogens, and its role in virus resistance remains unclear. Here, we cloned CaPR1 from the ChiVMV-highly resistant pepper variety ‘Perennial’. The 477 bp ORF encodes a 17.65 kDa basic protein containing a conserved CAP-PR1 domain. The subcellular localization of CaPR1 revealed that it was located in the plasma membrane, endoplasmic reticulum (ER), and nucleus. RT-qPCR revealed leaf-predominant expression, with earlier and stronger induction in the highly resistant than the highly susceptible variety after ChiVMV inoculation (6.4-fold at 2 days post-inoculation). The overexpression of CaPR1 in tobacco significantly increased resistance, reducing disease index by 25% and viral coat protein accumulation. Our findings identified CaPR1 as a positive regulator of ChiVMV resistance, providing a molecular target for pepper breeding. In addition, exogenous SA treatment increased the resistance of the highly susceptible cultivar ‘Guijiao 12’ to ChiVMV, and 0.25 mM had a greater effect.
Full article
(This article belongs to the Special Issue Emerging and Reemerging Plant Viruses in a Changing World)
►▼
Show Figures

Figure 1
Open AccessArticle
Bayesian Estimation of the True Prevalence of Caprine Arthritis Encephalitis in Hungarian Goat Herds
by
Krisztina Bárdos, Marietta Máté, Katalin Veres, Zsolt Lang, Giuseppe Bertoni, Carlos Eduardo Abril, Snorre Stuen, Saulius Petkevičius, Marcin Mickiewicz, Michał Czopowicz, Jarosław Kaba and László Ózsvári
Viruses 2025, 17(11), 1455; https://doi.org/10.3390/v17111455 - 31 Oct 2025
Abstract
Background: Caprine arthritis encephalitis (CAE) is a major viral disease of goats, caused by small ruminant lentivirus (SRLV), associated with chronic arthritis, mastitis, pneumonia, and encephalitis, leading to economic losses and reduced animal welfare. This study aimed to estimate the true prevalence of
[...] Read more.
Background: Caprine arthritis encephalitis (CAE) is a major viral disease of goats, caused by small ruminant lentivirus (SRLV), associated with chronic arthritis, mastitis, pneumonia, and encephalitis, leading to economic losses and reduced animal welfare. This study aimed to estimate the true prevalence of CAE in Hungarian goat herds, based on nationwide sampling and statistical modeling. Methods: Blood samples from 1218 goats in 53 herds were tested using ELISA, and true prevalence was estimated by Bayesian analysis. Results: The mean herd true prevalence (HTP) was 29.1% (95% CrI: 20.8–38.5%), while within the infected herds, the conditional within herd prevalence (CWHP) reached 58% ± 27.1%. Medium- and large-sized herds (>50 animals) showed the highest mean HTP (77.8% and 74.9%, respectively). No significant regional differences were observed, indicating that CAE is uniformly distributed across the country. Conclusions: Our findings place Hungary among moderately to highly affected European countries and highlight the need for a nationwide control strategy integrating routine serological surveillance, biosecurity improvements, farmer education, and long-term tools such as selective breeding.
Full article
(This article belongs to the Special Issue Viral Diseases of Sheep and Goats)
►▼
Show Figures

Figure 1

Journal Menu
► ▼ Journal Menu-
- Viruses Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics

Conferences
Special Issues
Special Issue in
Viruses
Opportunistic Viral Infections, 3rd Edition
Guest Editor: Carlo ContiniDeadline: 7 November 2025
Special Issue in
Viruses
Virus-Host Interactions: From Mechanisms to Therapeutics
Guest Editors: Leike Zhang, Ke XuDeadline: 15 November 2025
Special Issue in
Viruses
Recent Advances on Arboviruses Pathogenesis and Evolution
Guest Editor: Mariana Sequetin CunhaDeadline: 17 November 2025
Special Issue in
Viruses
Advances in Plant Virus/Viroid Detection and Identification Methods, 2nd Edition
Guest Editors: Laura Tomassoli, Antonio TiberiniDeadline: 28 November 2025
Topical Collections
Topical Collection in
Viruses
Efficacy and Safety of Antiviral Therapy
Collection Editors: Giordano Madeddu, Andrea De Vito, Agnese Colpani










