Journal Description
Genes
Genes
is a peer-reviewed, open access journal of genetics and genomics published monthly online by MDPI. The Spanish Society for Nitrogen Fixation (SEFIN) is affiliated with Genes and their members receive discounts on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, MEDLINE, PMC, Embase, PubAg, and other databases.
- Journal Rank: JCR - Q2 (Genetics and Heredity) / CiteScore - Q2 (Genetics (clinical))
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 14.6 days after submission; acceptance to publication is undertaken in 2.5 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: Reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Impact Factor:
2.8 (2024);
5-Year Impact Factor:
3.2 (2024)
Latest Articles
PTEN Gene and Autism: Genetic Underpinnings and Neurodevelopmental Impacts
Genes 2025, 16(9), 1061; https://doi.org/10.3390/genes16091061 - 9 Sep 2025
Abstract
Background/Objectives: Twin and family studies suggest that 90% of the risk for autism spectrum disorder (ASD) is due to genetic factors, with 800 genes recognized as playing a role. An important gene is phosphatase and tensin homolog (PTEN), which plays
[...] Read more.
Background/Objectives: Twin and family studies suggest that 90% of the risk for autism spectrum disorder (ASD) is due to genetic factors, with 800 genes recognized as playing a role. An important gene is phosphatase and tensin homolog (PTEN), which plays a significant role in cancer as a tumor suppressor best known for causing overgrowth and PTEN hamartoma tumor syndromes (PHTS). Less well known are PTEN germline mutations with adverse neurodevelopmental impacts of macrocephaly, intellectual disability, and ASD, as well as other behavioral and psychiatric disturbances. There remains a limited understanding of whether these gene variants are associated with differing manifestations of PTEN-associated neurodevelopmental disorders. Methods: This review utilized comprehensive literature searches such as PubMed, OMIM, and Gene Reviews with keywords of PTEN, genetic factors, autism, and human studies and by searching genomic-protein functional networks with STRING computer-based programs for functional and genetic mechanisms. Results: This review explored the genetic underpinnings of PTEN gene variants causing altered interactive proteins and their mechanisms, biological processes, molecular functions, pathways, and disease–gene associations. We characterized specific gene–gene or protein–protein interactions and their functions relating to neurodevelopment, psychiatric disorders, and ASD that were found to be increased with PTEN gene variants. Conclusions: PTEN gene defects are among the most recognized genetic causes of ASD. PTEN gene variants and altered protein interactions and mechanisms described in our study are associated with an increased risk for tissue and organ overgrowth, macrocephaly, and distinct brain anomalies, specifically newly identified abnormal CSF dynamics. These genetic underpinnings and impacts on neurodevelopment are discussed. The genetic and protein findings identified may offer clues to effective treatment interventions, particularly when instituted at a young age, to improve long-term outcomes.
Full article
(This article belongs to the Section Neurogenomics)
►
Show Figures
Open AccessReview
MicroRNA (miRNA) in the Pathogenesis of Diabetic Retinopathy: A Narrative Review
by
Stamatios Lampsas, Chrysa Agapitou, Alexandros Chatzirallis, Georgios Papavasileiou, Dimitrios Poulakis, Sofia Pegka, Panagiotis Theodossiadis, Vaia Lambadiari and Irini Chatziralli
Genes 2025, 16(9), 1060; https://doi.org/10.3390/genes16091060 - 9 Sep 2025
Abstract
Diabetic retinopathy (DR) is the most common microvascular complication associated with diabetes mellitus and represents a leading cause of visual impairment worldwide. Inflammation, endothelial dysfunction, angiogenesis, neurodegeneration, and oxidative stress are key pathogenic processes in the development and progression of DR. Numerous microRNAs
[...] Read more.
Diabetic retinopathy (DR) is the most common microvascular complication associated with diabetes mellitus and represents a leading cause of visual impairment worldwide. Inflammation, endothelial dysfunction, angiogenesis, neurodegeneration, and oxidative stress are key pathogenic processes in the development and progression of DR. Numerous microRNAs (miRNAs) show altered expression in DR and modulate critical biological pathways. Pro-inflammatory miRNAs such as miR-155 and miR-21 promote cytokine release and vascular inflammation, while miR-146a acts as a negative regulator of Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling. MiR-126 and miR-21 regulate endothelial integrity and angiogenesis through pathways involving Vascular Endothelial Growth Factor (VEGF). MiR-200b and miR-126 are downregulated in DR, leading to increased neovascularization via activation of the VEGF/ Mitogen-Activated Protein Kinase (MAPK) cascade. Apoptotic processes are affected by miR-195, which downregulates Sirtuin 1 (SIRT1) and B-cell lymphoma 2 (Bcl-2), promoting retinal cell death, while miR-29b downregulation permits upregulation of the transcription factor SP1, enhancing caspase-mediated apoptosis in Müller cells and endothelial cells. miRNAs collectively modulate an intricate regulatory network that contributes to the underlying mechanisms of diabetic retinopathy development and progression. This narrative review aims to summarize knowledge regarding the mechanisms miRNAs mediating pathogenetic mechanisms of DR.
Full article
(This article belongs to the Section RNA)
►▼
Show Figures

Figure 1
Open AccessArticle
Destructive and Non-Destructive Methods for aDNA Isolation from Teeth and Their Analysis: A Comparison
by
Agnieszka Dobosz, Anna Jonkisz, Arleta Lebioda, Jerzy Kawecki and Tadeusz Dobosz
Genes 2025, 16(9), 1059; https://doi.org/10.3390/genes16091059 - 9 Sep 2025
Abstract
Background/Objectives: DNA analysis can be used to expand our understanding of extinct populations and the history of the world and humankind. Dental cavities often contain uncontaminated remains of ancient DNA (aDNA). Archaeological excavations are a convenient source for various samples; however, in almost
[...] Read more.
Background/Objectives: DNA analysis can be used to expand our understanding of extinct populations and the history of the world and humankind. Dental cavities often contain uncontaminated remains of ancient DNA (aDNA). Archaeological excavations are a convenient source for various samples; however, in almost all extraction methods, a piece of bone or tooth is powdered before extraction, thereby causing damage to archaeological samples that are often irreplaceable and unique. This study aimed to develop a method that enables the collection of DNA from teeth without causing significant damage. Methods: This study presents two methods of DNA extraction from teeth: destructive and non-destructive. Both contemporary and archaeological teeth were examined using both destructive and non-destructive approaches to compare their efficiency. To verify the results, methods such as quantitative RT-PCR, STR analysis, and Y-SNP analysis were employed. Results: Extraction efficiency plays a critical role in this field of research. The main steps of the DNA extraction method were compared and optimized based on purification and using quantitative PCR. Conclusions: The results demonstrate that a non-destructive method of DNA isolation from human teeth can be used successfully, especially when teeth are unique and cannot be destroyed during the examination process. This method yields an appropriate amount of DNA for sequencing.
Full article
(This article belongs to the Section Technologies and Resources for Genetics)
►▼
Show Figures
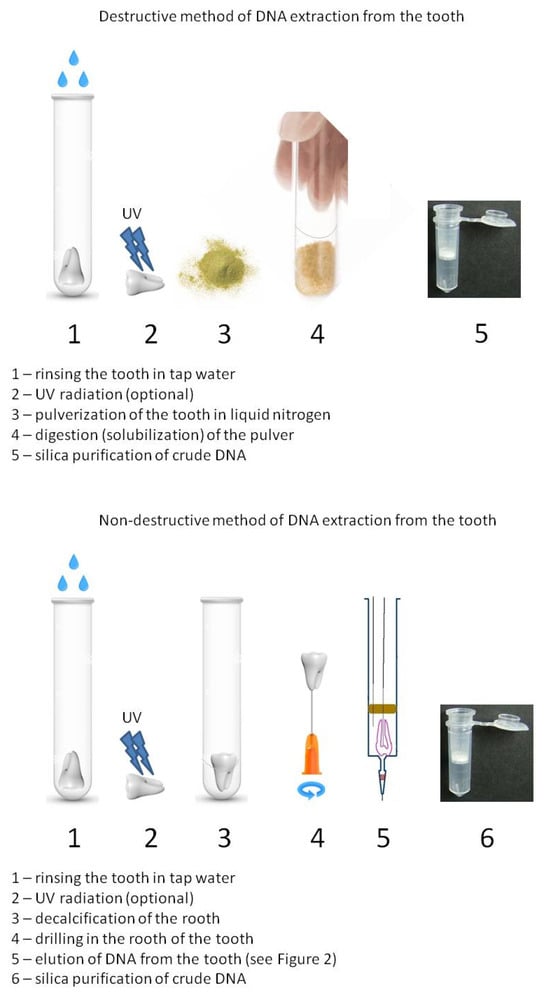
Figure 1
Open AccessReview
The Impact of Genetic Polymorphisms on the Clinical Efficacy of Azole Antifungals
by
Hareesh Singam and Sherif Mossad
Genes 2025, 16(9), 1058; https://doi.org/10.3390/genes16091058 - 9 Sep 2025
Abstract
Azoles are the primary agents for antifungal activity in clinical medicine due to their broad-spectrum efficacy and favorable safety profiles compared to older agents. Triazoles, including fluconazole, itraconazole, voriconazole, posaconazole, and isavuconazole, have varied pharmacokinetic and pharmacodynamic properties. This is due to various
[...] Read more.
Azoles are the primary agents for antifungal activity in clinical medicine due to their broad-spectrum efficacy and favorable safety profiles compared to older agents. Triazoles, including fluconazole, itraconazole, voriconazole, posaconazole, and isavuconazole, have varied pharmacokinetic and pharmacodynamic properties. This is due to various polymorphisms in hepatic enzymes, necessitating genotype-guided dosing and therapeutic drug monitoring (TDM) to optimize treatment outcomes. This review highlights the clinical relevance of pharmacogenomics in azole therapy, particularly the role of cytochrome P450 (CYP450) enzyme polymorphisms in influencing drug levels, efficacy, and toxicity. Understanding these genetic and metabolic factors is essential for personalized antifungal treatment strategies, improving patient safety and therapeutic outcomes.
Full article
(This article belongs to the Special Issue Progress in Hematology: Non-Malignant, Pre-Malignant, and Malignant Disorders, and Genetically Based Therapies)
Open AccessReview
Angiogenic microRNAs in Systemic Sclerosis: Insights into Microvascular Dysfunction and Therapeutic Implications
by
Marta Rusek
Genes 2025, 16(9), 1057; https://doi.org/10.3390/genes16091057 - 9 Sep 2025
Abstract
Systemic sclerosis (SSc) is a complex connective tissue disease that affects the skin and internal organs and is characterized by immune dysregulation, progressive fibrosis, and microvascular dysfunction. Chronic tissue ischemia, accompanied by impaired angiogenesis, leads to the gradual loss of small vessels, resulting
[...] Read more.
Systemic sclerosis (SSc) is a complex connective tissue disease that affects the skin and internal organs and is characterized by immune dysregulation, progressive fibrosis, and microvascular dysfunction. Chronic tissue ischemia, accompanied by impaired angiogenesis, leads to the gradual loss of small vessels, resulting in clinical complications, such as Raynaud’s phenomenon, digital ulcers, pulmonary arterial hypertension, and renal crisis. Emerging evidence highlights the crucial regulatory role of microRNAs (miRNAs) in vascular homeostasis through the modulation of key signaling pathways and endothelial cell activity. Dysregulated miRNAs influence fibroblast proliferation, inflammatory responses, and immune cell activity in SSc, contributing to disease progression. Current knowledge is still limited, highlighting the need for further research to elucidate the miRNAs network involved in the etiopathogenesis of SSc. The use of miRNA-based biomarkers is gaining tremendous attention for early diagnosis, risk stratification, classification, and the prediction of therapeutic responses. This review provides insights into angiogenesis-related miRNAs involved in SSc pathogenesis, discusses their relevance as biomarkers, and explores their promise as therapeutic targets. Advancing our knowledge of miRNAs-mediated regulatory networks may open new possibilities for personalized approaches to SSc management.
Full article
(This article belongs to the Section RNA)
►▼
Show Figures
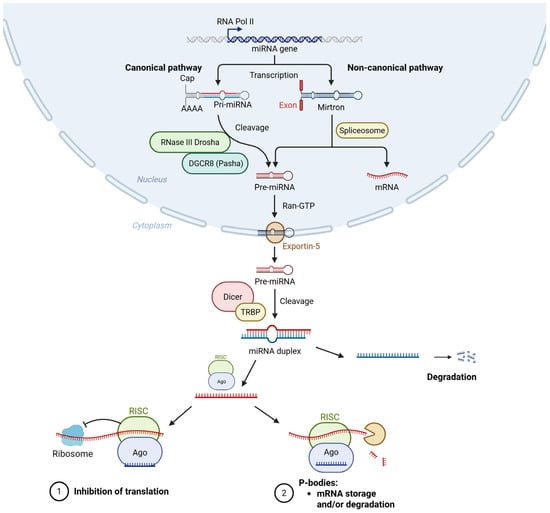
Figure 1
Open AccessArticle
Screening and Stability Analysis of Reference Genes in Pastor roseus
by
Xixiu Sun, Ran Li, Xiaojie Wang, Hongxia Hu, Kun Yang, Jianguo Wu, Jun Lin, Rong Ji and Xiaofang Ye
Genes 2025, 16(9), 1056; https://doi.org/10.3390/genes16091056 - 9 Sep 2025
Abstract
Background/Objectives: Optimal reference genes for normalizing RT-qPCR data depend on the species, treatments, developmental stages, and other conditions. Pastor roseus is a long-distance migratory bird with potential applications in locust biological control. This study applied reverse transcription quantitative PCR (RT-qPCR) to evaluate the
[...] Read more.
Background/Objectives: Optimal reference genes for normalizing RT-qPCR data depend on the species, treatments, developmental stages, and other conditions. Pastor roseus is a long-distance migratory bird with potential applications in locust biological control. This study applied reverse transcription quantitative PCR (RT-qPCR) to evaluate the expression stability of six genes (RPS2, ACTB, B2M, SDHA, UBE2G2, and RPL4) in blood samples from female, male, and nestling P. roseus. Methods: An integrated analysis of the expression stability of six reference genes was performed using three statistical algorithms: GeNorm, BestKeeper, and NormFinder. Results: The results showed that SDHA, ACTB, and B2M exhibited the highest expression stability among the candidate reference genes. The optimal number of reference genes was two, as determined by a pairwise variation analysis using GeNorm. Subsequent comprehensive validation using RefFinder identified SDHA/ACTB as the optimal reference gene pair for normalizing gene expression data for P. roseus. Conclusions: These findings establish a robust foundation for ensuring data accuracy in functional genomic studies of P. roseus.
Full article
(This article belongs to the Special Issue Genetic Breeding of Poultry)
►▼
Show Figures

Graphical abstract
Open AccessReview
A Comprehensive Analysis Examining the Role of Genetic Influences on Psychotropic Medication Response in Children
by
Jatinder Singh, Athina Manginas, Georgina Wilkins and Paramala Santosh
Genes 2025, 16(9), 1055; https://doi.org/10.3390/genes16091055 (registering DOI) - 8 Sep 2025
Abstract
Psychotropic medication is commonly used for the treatment of mental health conditions. However, the genetic factors that influence psychotropic medication responses in children have not been thoroughly investigated. To address this gap, a systematic review and thematic analysis were conducted to examine the
[...] Read more.
Psychotropic medication is commonly used for the treatment of mental health conditions. However, the genetic factors that influence psychotropic medication responses in children have not been thoroughly investigated. To address this gap, a systematic review and thematic analysis were conducted to examine the genetic impact of psychotropic medication response in children. The Down and Blacks and Consolidated Health Economic Evaluation Reporting Standards (CHEERS) checklists assessed the quality of studies and health economics, respectively. Using PRISMA reporting guidelines, 50 articles were identified with a sample size ranging from 2 to 2.9 million individuals. Most of the studies reported on ethnicity, and approximately half of the studies (24/50) were performed in North America. Five themes emerged from the thematic analysis: (1) implications of non-CYP450 polymorphisms, (2) paediatric CYP450 pharmacogenetics, (3) genetic predictors of response, (4) insights for implementation and future research and (5) phenoconversion. The thematic analysis revealed that assessment of non-CYP450 polymorphisms and psychotropic medication response, especially in those with mental health conditions such as autism, would be helpful. Epilepsy onset, risk and treatment response were associated with non-CYP450 genetic variants. Phenoconversion of substrates associated with CYP2D6 and CYP2C19 metabolisers is common in individuals with mental health conditions, and ABCB1 variants can influence psychotropic medication responses. A multidisciplinary model could also help guide clinical decision-making in cases involving complex neurodevelopmental profiles. Using the Down and Blacks checklist, the average score from the 50 studies was 17.7 points (min. 14, max. 24). The health economic evaluation of studies using the CHEERS checklist gave an average score of 33.0% (range: 21.4% to 35.7%). The study provides an important resource of information for healthcare professionals, researchers and policymakers working at the intersection of child psychiatry, pharmacogenomics and precision medicine.
Full article
(This article belongs to the Section Neurogenomics)
►▼
Show Figures
Figure 1
Open AccessArticle
Insights into Genomic Patterns of Homozygosity in the Endangered Dülmen Wild Horse Population
by
Silke Duderstadt and Ottmar Distl
Genes 2025, 16(9), 1054; https://doi.org/10.3390/genes16091054 - 8 Sep 2025
Abstract
Background/Objectives: Dülmen wild horses are kept in a fenced wooden and marsh area around Dülmen in Westphalia, Germany, since 1856. Previous analyses supported early genetic divergence from other domesticated horse populations and the Przewalski horse. Therefore, the objective of this study was to
[...] Read more.
Background/Objectives: Dülmen wild horses are kept in a fenced wooden and marsh area around Dülmen in Westphalia, Germany, since 1856. Previous analyses supported early genetic divergence from other domesticated horse populations and the Przewalski horse. Therefore, the objective of this study was to evaluate genetic diversity using high-density genomic data. Methods: We collected 337 one-year-old male Dülmen wild horses, captured at 12 annual auctions, for genotyping on the Illumina GGP Equine Plus Beadchip. All analyses were performed for 63,123 autosomal SNPs. Results: On average, each horse had 27.96 ROH with an average length of 8.237 Mb, resulting in an average genomic inbreeding coefficient FROH of 0.107. ROH with a length of 2–4 Mb were most frequent, and the next frequent ROH fall into the length categories of 4–8 and 8–16 Mb. The effective population size (Ne) steadily decreased in the last 100 generations by 4.57 individuals per generation from 498 to 41. We identified 10 ROH islands on equine chromosomes 1, 4, 5, 7, 9, and 10. Only one ROH island on ECA 1 was shared by 45% of the horses. Overrepresented genes of ROH islands were associated with glycerophospholipid catabolism through phospholipase A2 genes, skeletal muscle contraction (TNNI3, TNNT1), synapse activity and structure (CTTNBP2), regulation of inflammatory response (NLRP genes), and zinc finger protein genes, which are involved in many cellular processes and may also act as tumor suppressors and oncogenes. Conclusions: This study highlights the development of genomic inbreeding and shows the importance of the stallions selected for breeding on the genetic diversity of the Dülmen wild horses. The results of this study should be used to develop strategies to slow down increase in inbreeding and prevent transmitting unfavorable alleles from the stallions to the next generation.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
Open AccessArticle
Stutter Modeling in Probabilistic Genotyping for Forensic DNA Analysis: A Casework-Driven Assessment
by
Camila Costa, Érica Pereira, Sandra Costa, Paulo Miguel Ferreira, António Amorim, Lourdes Prieto and Nádia Pinto
Genes 2025, 16(9), 1053; https://doi.org/10.3390/genes16091053 - 8 Sep 2025
Abstract
Background: Probabilistic genotyping software has become an essential tool in forensic genetics, particularly for interpreting complex DNA mixtures. Previous studies measured the impact of considering widely divergent statistical approaches in quantifying evidence, both inter- and intra-software. At a much smaller scale, this data-driven
[...] Read more.
Background: Probabilistic genotyping software has become an essential tool in forensic genetics, particularly for interpreting complex DNA mixtures. Previous studies measured the impact of considering widely divergent statistical approaches in quantifying evidence, both inter- and intra-software. At a much smaller scale, this data-driven study shows how different models implemented on distinct versions of the same tool may affect the results. Among the available tools, EuroForMix stands out as a quantitative, open-source software that models various aspects of the DNA profile, including artefacts like stutter peaks. Its freeware nature allowed the use of both versions 1.9.3. and 3.4.0, between which several updates were made, including the possibility to model both back and forward stutter, compared to only modeling back stutters inputted by the expert in the earlier version. Methods: A total of 156 real casework sample pairs (comprising mixtures with two or three estimated contributors and associated reference) from the Portuguese Scientific Police Laboratory were analyzed using both software versions. The same input data, containing alleles and artefactual peaks, were used to reflect operational conditions. Statistical measurements were compared and further investigated. Results: Most Likelihood Ratio values differed in less than one order of magnitude across versions. However, exceptions were found in more complex samples, such as those with more contributors, unbalanced contributions, or greater degradation. Conclusions: This work emphasizes the relevance of model selection in forensic evidence quantification, even when considering different versions of the same tool. The impact of different models in statistical evaluation depends on several factors, such as sample technical conditions, genotypic profiles, and population distribution.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures
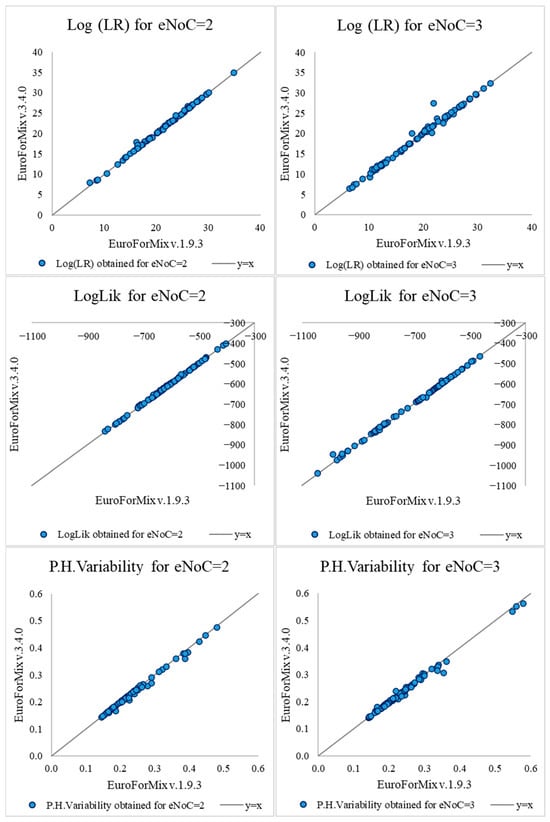
Figure 1
Open AccessArticle
Detection of Clinically Significant BRCA Large Genomic Rearrangements in FFPE Ovarian Cancer Samples: A Comparative NGS Study
by
Alessia Perrucci, Maria De Bonis, Giulia Maneri, Claudio Ricciardi Tenore, Paola Concolino, Matteo Corsi, Alessandra Conca, Jessica Evangelista, Alessia Piermattei, Camilla Nero, Luciano Giacò, Elisa De Paolis, Anna Fagotti and Angelo Minucci
Genes 2025, 16(9), 1052; https://doi.org/10.3390/genes16091052 - 8 Sep 2025
Abstract
Background: Copy number variations (CNVs), also referred to as large genomic rearrangements (LGRs), represent a crucial component of BRCA1/2 (BRCA) testing. Next-generation sequencing (NGS) has become an established approach for detecting LGRs by combining sequencing data with dedicated bioinformatics pipelines. However, CNV detection
[...] Read more.
Background: Copy number variations (CNVs), also referred to as large genomic rearrangements (LGRs), represent a crucial component of BRCA1/2 (BRCA) testing. Next-generation sequencing (NGS) has become an established approach for detecting LGRs by combining sequencing data with dedicated bioinformatics pipelines. However, CNV detection in formalin-fixed paraffin-embedded (FFPE) samples remains technically challenging, and there is the need to implement a robust and optimized analysis strategy for routine clinical practice. Methods: This study evaluated 40 FFPE ovarian cancer (OC) samples from patients undergoing BRCA testing. The performance of the amplicon-based NGS Diatech Myriapod® NGS BRCA1/2 panel (Diatech Pharmacogenetics, Jesi, Italy) was assessed for its ability to detect BRCA CNVs and results were compared to two hybrid capture-based reference assays. Results: Among the 40 analyzed samples (17 CNV-positive and 23 CNV-negative for BRCA genes), the Diatech pipeline showed a good concordance with the reference method—all CNVs were correctly identified in 16 cases with good enough sequencing quality. Only one result was inconclusive due to low sequencing quality. Conclusions: These findings support the clinical utility of NGS-based CNV analysis in FFPE samples when combined with appropriate bioinformatics tools. Integrating visual inspection of CNV plots with automated CNV calling improves the reliability of CNV detection and enhances the interpretation of results from tumor tissue. Accurate CNV detection directly from tumor tissue may reduce the need for additional germline testing, thus shortening turnaround times. Nevertheless, blood-based testing remains mandatory to determine whether detected BRCA CNVs are of hereditary or somatic origin, particularly in cases with a strong clinical suspicion of inherited predisposition due to young age and a personal and/or family history of OC.
Full article
(This article belongs to the Section Human Genomics and Genetic Diseases)
►▼
Show Figures

Figure 1
Open AccessArticle
Comparative Genomics and Functional Profiling Reveal Lineage-Specific Metabolic Adaptations in Globally Emerging Fluoroquinolone-Resistant Salmonella Kentucky ST198
by
Juned Ahmed, Rachel C. Soltys, Smriti Shringi, Jean Guard, Bradd J. Haley and Devendra H. Shah
Genes 2025, 16(9), 1051; https://doi.org/10.3390/genes16091051 - 8 Sep 2025
Abstract
Background: Salmonella Kentucky comprises two major lineages, ST152 and fluoroquinolone-resistant (FluR) ST198, which have diverged genotypically and phenotypically along distinct evolutionary and epidemiological trajectories. ST198 is linked to global human disease, while ST152 is primarily animal-associated in the U.S. We hypothesized
[...] Read more.
Background: Salmonella Kentucky comprises two major lineages, ST152 and fluoroquinolone-resistant (FluR) ST198, which have diverged genotypically and phenotypically along distinct evolutionary and epidemiological trajectories. ST198 is linked to global human disease, while ST152 is primarily animal-associated in the U.S. We hypothesized that lineage-specific metabolic adaptations contribute to their differing host associations and pathogenicity. Methods: We performed comparative metabolic profiling of ST198 (n = 3) and ST152 (n = 4) strains across 948 substrates and environmental conditions. Growth assays tested the ability of these lineages and other non-typhoidal Salmonella (NTS) serovars (n = 5) to utilize myo-inositol and lactulose as sole carbon sources. Comparative genomic analyses of 294 ST198, 173 ST152, and 1300 other NTS serovars identified nutrient utilization genes. Results: ST198 exhibited significantly higher respiratory activity and broader metabolic versatility across carbon, nitrogen/sulfur sources, and stress conditions. The canonical iol gene cluster for myo-inositol catabolism was conserved in ST198 but absent in ST152, which nonetheless showed weak growth on myo-inositol, suggesting an alternative metabolic pathway for myo-inositol may exist. We also report for the first time that, despite lineage-specific differences in metabolic efficiency, multiple NTS serovars, including S. Kentucky, can metabolize lactulose, a synthetic disaccharide traditionally associated with beneficial gut microbes. These results suggest the potential existence of a novel lactulose metabolic pathway in NTS. Conclusions: These findings highlight ST198’s metabolic adaptability and reveal novel metabolic capacities in NTS. A mechanistic understanding of nutrient utilization pathways, particularly of myo-inositol and lactulose, will provide novel insights into the mechanisms underlying nutrient metabolism that likely modulate the ecological success and pathogenic potential of NTS in human and animal hosts.
Full article
(This article belongs to the Special Issue Advances in Molecular Microbiology, Genetics, and Bioinformatics of Multiple-Drug-Resistant Bacteria in Public Health)
►▼
Show Figures

Figure 1
Open AccessArticle
Genetic Mapping and Diversity of Indigenous and Exotic Rabbits: Adaptive and Conservation Strategies
by
Marwa M. Ahmed, Shaymaa M. Abousaad, Soha S. Abdel-Magid, Shoukry M. El-Tantawi, Hatem M. Ali, Essam A. El-Gendy, Nour A. Abouzeid, Lin Yang, Kaliyah Hayes, Mackenzie Skye. Hamilton, Ayman M. Abouzeid and Yongjie Wang
Genes 2025, 16(9), 1050; https://doi.org/10.3390/genes16091050 - 8 Sep 2025
Abstract
Background: Climate change threatens global food security, highlighting the need for adaptive traits in livestock to ensure sustainable production. Rabbits, known for their unique adaptability, require the preservation of genetic diversity to maintain resilience. The decline in genetic specificity among indigenous breeds underscores
[...] Read more.
Background: Climate change threatens global food security, highlighting the need for adaptive traits in livestock to ensure sustainable production. Rabbits, known for their unique adaptability, require the preservation of genetic diversity to maintain resilience. The decline in genetic specificity among indigenous breeds underscores the urgency of conservation efforts to protect these critical resources. Objectives: This study investigates the genetic structure and diversity of indigenous rabbit populations, emphasizing genetic mapping as essential for sustaining adaptability. The findings aim to guide breeding programs that enhance biodiversity and support agricultural resilience. Materials and Methods: This study analyzed both native and exotic rabbit breeds. Native breeds included Black Baladi (BB), White Baladi (WB), Red Baladi (RB), and Jabali (JAB), while exotic breeds included New Zealand White (NZW), American Rex (AR), and Chinchilla (CH). Fourteen microsatellite loci were genotyped in 526 rabbits across all breeds. Results: A total of 467 alleles were identified, with an overall mean of 5.03. The expected heterozygote frequencies were medium to high. Polymorphism was high in BB, JAB, and NZW, and medium in WB, RB, AR, and CH. FIS and FIT values (−0.044 and 0.156) suggested possible non-intensive inbreeding. FST (0.220) showed breed differentiation and high within-breed variation. The gene flow averaged 1.872, indicating interbreed gene exchange. Neutrality and phylogenetic analyses revealed genetic reshaping; BB, WB, RB, AR, CH, and NZW showed overlap, while JAB retained high specificity. Conclusions: Urgent conservation strategies are essential to preserve native rabbit genetic diversity and unique traits, which are vital for sustaining biodiversity and livestock resilience globally.
Full article
(This article belongs to the Section Animal Genetics and Genomics)
►▼
Show Figures
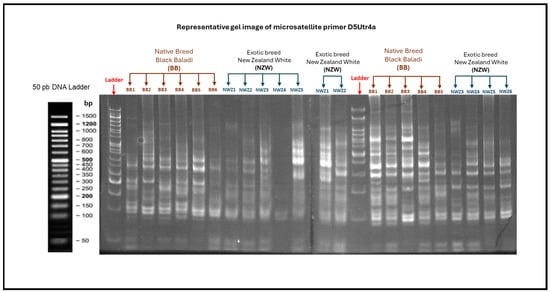
Figure 1
Open AccessReview
Gut Dysbiosis Driven by CFTR Gene Mutations in Cystic Fibrosis Patients: From Genetic Disruption to Multisystem Consequences and Microbiota Modulation
by
Natalia Pawłowska, Magdalena Durda-Masny, Szczepan Cofta, Daria Springer and Anita Szwed
Genes 2025, 16(9), 1049; https://doi.org/10.3390/genes16091049 - 6 Sep 2025
Abstract
Mutations in the CFTR genes causing cystic fibrosis (CF) are associated with the presence of thick, viscous mucus and the formation of biofilms in the gastrointestinal tract (GI) that impair intestinal homeostasis, triggering chronic inflammation, epithelial barrier dysfunction, and changes in the composition
[...] Read more.
Mutations in the CFTR genes causing cystic fibrosis (CF) are associated with the presence of thick, viscous mucus and the formation of biofilms in the gastrointestinal tract (GI) that impair intestinal homeostasis, triggering chronic inflammation, epithelial barrier dysfunction, and changes in the composition and activity of the gut microbiota. CFTR protein modulators represent a promising approach to enhancing lower GI function in patients with CF. The aim of the review is to present the complex relationships between the presence of CFTR gene mutations and the gut microbiota dysbiosis in patients with cystic fibrosis. Mutations in the CFTR gene, the molecular basis of cystic fibrosis (CF), disrupt epithelial ion transport and profoundly alter the gastrointestinal environment. Defective chloride and bicarbonate secretion leads to dehydration of the mucosal layer, increased mucus viscosity, and the formation of biofilms that favour microbial persistence, which together promote gut microbiota dysbiosis. This dysbiotic state contributes to impaired epithelial barrier function, chronic intestinal inflammation, and abnormal immune activation, thereby reinforcing disease progression. The interplay between CFTR dysfunction and microbial imbalance appears to be bidirectional, as dysbiosis may further exacerbate epithelial stress and inflammatory signalling. Therapeutic interventions with CFTR protein modulators offer the potential to partially restore epithelial physiology, improve mucus hydration, and foster a microbial milieu more consistent with intestinal homeostasis. The aim of this review is to elucidate the complex relationships between CFTR gene mutations and gut microbiota dysbiosis in patients with cystic fibrosis, with a particular emphasis on the clinical implications of these interactions and their potential to inform novel therapeutic strategies.
Full article
(This article belongs to the Section Microbial Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
A Comprehensive Analysis of Transcriptomics and Proteomics Elucidates the Cold-Adaptive Ovarian Development of Eriocheir sinensis Farmed in High-Altitude Karst Landform
by
Qing Li, Yizhong Zhang and Lijuan Li
Genes 2025, 16(9), 1048; https://doi.org/10.3390/genes16091048 - 6 Sep 2025
Abstract
Background: In high-altitude regions, sporadic two-year-old immature Chinese mitten crabs (Eriocheir sinensis) would overwinter and mature in their third year, developing into three-year-old crabs (THCs) with a cold-adaptive strategy. Compared to two-year-old crabs (TWCs) from low-altitude Jiangsu, THCs from Karst landform
[...] Read more.
Background: In high-altitude regions, sporadic two-year-old immature Chinese mitten crabs (Eriocheir sinensis) would overwinter and mature in their third year, developing into three-year-old crabs (THCs) with a cold-adaptive strategy. Compared to two-year-old crabs (TWCs) from low-altitude Jiangsu, THCs from Karst landform and high-altitude Guizhou exhibit significantly larger final size but lower gonadosomatic index (GSI) (p < 0.01). Methods: To elucidate the molecular mechanisms underlying this delayed ovarian development, integrated transcriptomic and proteomic analyses were conducted. Results: Results showed downregulation of PI3K-Akt and FoxO signaling pathways, as well as upregulation of protein digestion and absorption pathways. Differentially expressed proteins indicated alterations in mitochondrial energy transduction and nutrient assimilation. Integrated omics analysis revealed significant changes in nucleic acid metabolism, proteostasis, and stress response, indicating systemic reorganization in energy-nutrient coordination and developmental plasticity. Conclusions: The observed growth-reproductive inverse relationship reflects an adaptive life-history trade-off under chronic cold stress, whereby energy repartitioning prioritizes somatic growth over gonadal investment. Our transcriptomic and proteomic data further suggest a pivotal regulatory role for FOXO3 dephosphorylation in potentially coupling altered energy sensing to reproductive suppression. This inferred mechanism reveals a potential conserved pathway for environmental adaptation in crustaceans, warranting further functional validation.
Full article
(This article belongs to the Section Molecular Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
Multi-SNP Haplotypes in Circadian PER3 Gene Are Associated with Mood and Sleep Disorders in University Students
by
Francesca Goodell and Krista K. Ingram
Genes 2025, 16(9), 1047; https://doi.org/10.3390/genes16091047 - 5 Sep 2025
Abstract
Background: Mood disorders, including anxiety, depression, and seasonal affective disorder (SAD), are often comorbid and can be exacerbated by the misalignment of an individual’s circadian rhythm with their social timing. Single-nucleotide polymorphisms (SNPs) in circadian clock genes have been associated with both
[...] Read more.
Background: Mood disorders, including anxiety, depression, and seasonal affective disorder (SAD), are often comorbid and can be exacerbated by the misalignment of an individual’s circadian rhythm with their social timing. Single-nucleotide polymorphisms (SNPs) in circadian clock genes have been associated with both internalizing disorders and sleep disturbances, and some clock polymorphisms, including those in the Period3 (PER3) gene, likely function via delaying or advancing circadian period and affecting sleep–wake patterns. Methods: Here, we explore associations of multiple SNP haplotypes in the PER3 gene with anxiety, depression, internalizing disorder (ID), chronotype, and sleep disturbance in young adults (n = 1109 individuals). Results: We report novel, sex-specific associations of single PER3 SNPs with mood and sleep disorders and highlight strong multi-SNP haplotype associations, revealing a greater risk of mood and sleep disorders in university students with specific PER3 haplotypes. Conclusions: Our results suggest that the additive effects of multiple risk variants amplify the prevalence of mood disorders and sleep disruptions in young adults. Understanding how polymorphisms within circadian genes interact to alter clock function, sleep-wake behavior and downstream physiological changes in the brain may help explain the comorbidity of mood and sleep syndromes and provide future therapeutic targets to combat these debilitating disorders.
Full article
(This article belongs to the Special Issue Genetics of Neuropsychiatric Disorders)
►▼
Show Figures
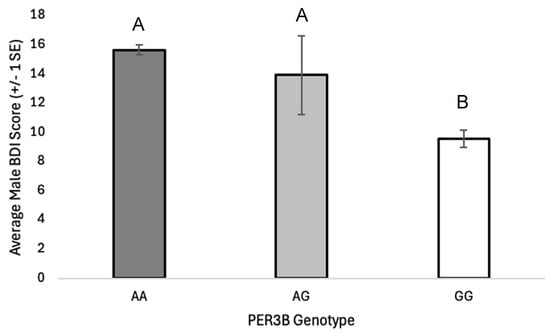
Figure 1
Open AccessArticle
Identification and Functional Speculation of Genes Related to Sex Pheromone Synthesis Expressed in the Gonads of Female Gynaephora qinghaiensis (Lepidoptera: Lymantriidae)
by
Zhanling Liu, Shujing Gao, Haibin Han, Xiaorui Wang, Guixiang Kou, Haishun Wang and Yuantao Zhou
Genes 2025, 16(9), 1046; https://doi.org/10.3390/genes16091046 - 5 Sep 2025
Abstract
Background: Grassland desertification has garnered significant attention as a pressing issue. Among the key pests affecting plateau meadows, the Gynaephora qinghaiensis (Lepidoptera: Lymantriidae) poses a substantial threat in the Qinghai-Tibet Plateau region, highlighting the urgent need for effective, environmentally friendly control strategies. Insect
[...] Read more.
Background: Grassland desertification has garnered significant attention as a pressing issue. Among the key pests affecting plateau meadows, the Gynaephora qinghaiensis (Lepidoptera: Lymantriidae) poses a substantial threat in the Qinghai-Tibet Plateau region, highlighting the urgent need for effective, environmentally friendly control strategies. Insect sex pheromones are increasingly employed in pest monitoring and management. Methods: This study aims to identify and analyze genes associated with sex pheromone synthesis in grassland caterpillars through transcriptome sequencing and tissue-specific expression analysis. Results: A total of 139,599 transcripts and 56,403 Unigenes were obtained from the sex pheromone glands transcriptome database. A total of 31 genes related to sex pheromone synthesis were identified, including 1 ACC, 8 DES, 6 AR, 7 FAR, 5 FAS, and 4 ACT genes. The expression levels of these genes varied significantly across different tissues in both male and female caterpillars (p < 0.05). GqinACC1, GqinDES1, GqinDES4, GqinDES8, GqinAR3, GqinFAR6, GqinACT2, and GqinACT3 exhibited significantly higher expression levels in the female gonads compared to other tissues (p < 0.01). Conclusions: We hypothesize that specific genes play specific roles in the pheromone synthesis pathways of pests, Key genes were identified based on expression patterns for subsequent functional studies. The results of this study offer valuable data support for subsequent investigations into the mechanisms underlying sex pheromone synthesis in G. qinghaiensis. Additionally, these findings may identify potential targets for future research on genes associated with pheromone biosynthesis, which could disrupt their chemical communication and contribute to grassland conservation efforts.
Full article
(This article belongs to the Special Issue 15th Anniversary of Genes: Feature Papers in the “Molecular Genetics and Genomics” Section)
►▼
Show Figures

Figure 1
Open AccessArticle
Teashirt and C-Terminal Binding Protein Interact to Regulate Drosophila Eye Development
by
Surya Jyoti Banerjee, Jennifer Curtiss, Chase Drucker and Harley Hines
Genes 2025, 16(9), 1045; https://doi.org/10.3390/genes16091045 - 5 Sep 2025
Abstract
Background and Objectives: The Drosophila retinal determination network comprises the transcription factor Teashirt (Tsh) and the transcription co-regulator C-terminal Binding Protein (CtBP), both of which are essential for normal adult eye development. Both Tsh and CtBP show a pattern of co-expression in
[...] Read more.
Background and Objectives: The Drosophila retinal determination network comprises the transcription factor Teashirt (Tsh) and the transcription co-regulator C-terminal Binding Protein (CtBP), both of which are essential for normal adult eye development. Both Tsh and CtBP show a pattern of co-expression in the proliferating cells anterior to the morphogenetic furrow that demarcates the boundary between the anteriorly placed proliferating eye precursor cells and the posteriorly placed differentiating photoreceptor cells in the larval eye-precursor tissue, the eye–antennal disc. The disc ultimately develops into the adult compound eyes, antenna, and other head structures. Both Tsh and CtBP were found to interact genetically during ectopic eye formation in Drosophila, and both were present in molecular complexes purified from gut and cultured cells. However, it remained unknown whether Tsh and CtBP molecules could interact in the eye–antennal discs and elicit an effect on eye development. The present study answers these questions. Methods: 5′ GFP-tagging of the tsh gene in the Drosophila genome and 5′ FLAG-tagging of the ctbp gene were accomplished by the CRISPR-Cas9 and BAC recombineering methods, respectively, to produce GFP-Tsh- and FLAG-CtBP-fused proteins in specific transgenic Drosophila strains. Verification of these proteins’ expression in the larval eye–antennal discs was performed by immunohistological staining and confocal microscopy. Genetic screening was performed to establish functional interaction between Tsh and CtBP during eye development. Scanning Electron Microscopy was performed to image the adult eye structure. Co-immunoprecipitation and GST pulldown assays were performed to show that Tsh and CtBP interact in the cells of the third instar eye–antennal discs. Results: This study reveals that Tsh and CtBP interact genetically and physically in the Drosophila third instar larval eye–antennal disc to regulate adult eye development. This interaction is likely to limit the population of the eye precursor cells in the larval eye disc of Drosophila. Conclusions: The relative abundance of Tsh and CtBP in the third instar larval eye–antennal disc can dictate the outcome of their interaction on the Drosophila eye formation.
Full article
(This article belongs to the Special Issue Genetics and Genomics of Retinal Development and Diseases)
►▼
Show Figures

Figure 1
Open AccessArticle
High-Mobility Group Box Protein 3 (HMGB3) Facilitates DNA Interstrand Crosslink Processing and Double-Strand Break Repair in Human Cells
by
Jillian Dangerfield, Anirban Mukherjee, Wade Reh, Anna Battenhouse and Karen M. Vasquez
Genes 2025, 16(9), 1044; https://doi.org/10.3390/genes16091044 - 4 Sep 2025
Abstract
Background/Objectives: DNA-damaging agents can contribute to genetic instability, and such agents are often used in cancer chemotherapeutic regimens due to their cytotoxicity. Thus, understanding the mechanisms involved in DNA damage processing can not only enhance our knowledge of basic DNA repair mechanisms
[...] Read more.
Background/Objectives: DNA-damaging agents can contribute to genetic instability, and such agents are often used in cancer chemotherapeutic regimens due to their cytotoxicity. Thus, understanding the mechanisms involved in DNA damage processing can not only enhance our knowledge of basic DNA repair mechanisms but may also be used to develop improved chemotherapeutic strategies to treat cancer. The high-mobility group box protein 1 (HMGB1) is a known nucleotide excision repair (NER) cofactor, and its family member HMGB3 has been implicated in chemoresistance in ovarian cancer. Here, we aim to understand the potential role(s) of HMGB3 in processing DNA damage. Methods: A potential role in NER was investigated using HMGB3 knockout human cell lines in response to UV damage. Subsequently, potential roles in DNA interstrand crosslink (ICL) and DNA double-strand break (DSB) repair were investigated using mutagenesis assays, metaphase spreads, foci formation, a variety of DNA repair assays, and TagSeq analyses in human cells. Results: Interestingly, unlike HMGB1, HMGB3 does not appear to play a role in NER. We found evidence to suggest that HMGB3 is involved in the processing of both DSBs and ICLs in human cells. Conclusions: These novel results elucidate a role for HMGB3 in DNA damage repair and, surprisingly, also indicate a distinct role of HMGB3 in DNA damage repair from that of HMGB1. These findings advance our understanding of the role of HMGB3 in chemotherapeutic drug resistance and as a target for new chemotherapeutic strategies in the treatment of cancer.
Full article
(This article belongs to the Special Issue DNA Repair, Genomic Instability and Cancer)
►▼
Show Figures

Figure 1
Open AccessArticle
Unraveling the Complex Physiological, Biochemical, and Transcriptomic Responses of Pea Sprouts to Salinity Stress
by
Xiaoyu Xie, Liqing Zhan, Xiuxiu Su and Tingqin Wang
Genes 2025, 16(9), 1043; https://doi.org/10.3390/genes16091043 - 3 Sep 2025
Abstract
Background: The escalating global salinization poses a significant threat to agricultural productivity, necessitating a thorough understanding of plant responses to high salinity. Pea sprouts (Pisum sativum), a nutrient-rich crop increasingly cultivated in salinized regions, serve as an ideal model for
[...] Read more.
Background: The escalating global salinization poses a significant threat to agricultural productivity, necessitating a thorough understanding of plant responses to high salinity. Pea sprouts (Pisum sativum), a nutrient-rich crop increasingly cultivated in salinized regions, serve as an ideal model for such investigations due to their rapid growth cycle and documented sensitivity to ionic stress. Methods: In order to understand the response of pea sprouts in physiological regulation, redox-metabolic adjustments, and transcriptome reprogramming under salt stress, we investigated the effects of high salt concentrations on the ascorbic acid–glutathione cycle, endogenous hormone levels, metabolite profiles, and gene expression patterns in it. Results: Our findings reveal early-phase antioxidant/hormonal adjustments, mid-phase metabolic shifts, and late-phase transcriptomic reprogramming of pea sprouts under salt conditions. In addition, a biphasic response in the ascorbic acid cycle was found, with initial increases in enzyme activities followed by a decline, suggesting a temporary enhancement of antioxidant defenses. Hormonal profiling indicated a significant increase in abscisic acid (ABA) and jasmonic acid (JA), paralleled by a decrease in indole acetic acid (IAA) and dihydrozeatin (DZ), underscoring the role of hormonal regulation in stress adaptation. Metabolomic analysis uncovered salt-induced perturbations in sugars, amino acids, and organic acids, reflecting the metabolic reconfiguration necessary for osmotic adjustment and energy reallocation. Transcriptomic analysis identified 6219 differentially expressed genes (DEGs), with a focus on photosynthesis, hormone signaling, and stress-responsive pathways, providing insights into the molecular underpinnings of salt tolerance. Conclusions: This comprehensive study offers novel insights into the complex mechanisms employed by pea sprouts to combat salinity stress, contributing to the understanding of plant salt tolerance and potentially guiding the development of salt-resistant crop varieties.
Full article
(This article belongs to the Section Plant Genetics and Genomics)
►▼
Show Figures

Figure 1
Open AccessArticle
Detection of Development-Specific MicroRNAs in Rabbit Embryos and Culture Media: A Potential Biomarker Approach for Embryo Quality Assessment
by
María Salinas, Nikolett Tokodyné Szabadi, Gréta Dévai, Martin Urbán, Arnold Tóth, Bence Lázár, Timea Pintér, Annamária Nemes, Péter Fancsovits, Lilla Bodrogi and Elen Gócza
Genes 2025, 16(9), 1042; https://doi.org/10.3390/genes16091042 - 3 Sep 2025
Abstract
MicroRNAs (miRNAs) are short, non-coding RNA molecules that play a crucial role in regulating various biological processes by influencing post-transcriptional gene expression and gene silencing. Background/Objectives: In this study, rabbit embryos were utilised as a model system to investigate potential biomarkers relevant
[...] Read more.
MicroRNAs (miRNAs) are short, non-coding RNA molecules that play a crucial role in regulating various biological processes by influencing post-transcriptional gene expression and gene silencing. Background/Objectives: In this study, rabbit embryos were utilised as a model system to investigate potential biomarkers relevant to human embryo development. Seven microRNAs (miRNAs) identified in the embryo culture medium were evaluated as biomarkers by analysing the correlation between their expression levels and the developmental quality of rabbit embryos at days 4 and 6. Methods: We analysed the expression of seven development-specific miRNAs (miR-24-3p, miR-28-3p, miR-103a-3p, miR-181a-5p, miR-191-5p, miR-320a-3p, miR-378a-3p) in 4-day-old and 6-day-old rabbit embryos, along with their culture media. Results: Our findings revealed significant differences in the expression levels of these miRNAs between the 4-day-old and 6-day-old embryos. On the other hand, the expression patterns observed in the culture medium samples showed less variation between the two age groups. Nonetheless, analysis of miRNA expression profiles in the spent culture medium from individually cultured embryos enabled the identification of lower-quality embryos, characterised by smaller size and impaired or delayed development. Conclusions: The detection of these miRNAs in embryo culture medium may serve as a reliable indicator of successful progression to the blastocyst stage. Our experimental results identified specific miRNAs whose expression profiles differ according to embryonic stage and quality, thereby reflecting key developmental milestones. Notably, the detectability of these miRNAs in the medium—without prior RNA isolation—indicates their active secretion into the extracellular environment. By synthesising our findings with the existing literature, we refined a panel of miRNAs essential for the development of implantation-competent embryos in both rabbits and humans. Consequently, we developed a non-invasive assay for predicting implantation and pregnancy outcomes, which may have significant applications in human reproductive medicine.
Full article
(This article belongs to the Section RNA)
►▼
Show Figures

Figure 1

Journal Menu
► ▼ Journal Menu-
- Genes Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Editor’s Choice Articles
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Editorial Office
Journal Browser
► ▼ Journal BrowserHighly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Biomolecules, IJMS, Marine Drugs, Molecules, Sci. Pharm., Genes, Pharmaceutics, Crystals
Bioinformatics in Drug Design and Discovery—2nd Edition
Topic Editors: Bing Niu, Suren Rao Sooranna, Pufeng DuDeadline: 30 September 2025
Topic in
Agriculture, Agronomy, Crops, Genes, Plants, DNA
Vegetable Breeding, Genetics and Genomics, 2nd Volume
Topic Editors: Umesh K. Reddy, Padma Nimmakayala, Georgia NtatsiDeadline: 31 October 2025
Topic in
Brain Sciences, CIMB, Epigenomes, Genes, IJMS, DNA
Genetics and Epigenetics of Substance Use Disorders
Topic Editors: Aleksandra Suchanecka, Anna Maria Grzywacz, Kszysztof ChmielowiecDeadline: 15 November 2025
Topic in
Animals, CIMB, Genes, IJMS, DNA
Advances in Molecular Genetics and Breeding of Cattle, Sheep, and Goats
Topic Editors: Xiukai Cao, Hui Li, Huitong ZhouDeadline: 30 November 2025

Special Issues
Special Issue in
Genes
Molecular Genetics of Rare Disorders
Guest Editor: Miriam Lucia CarrieroDeadline: 10 September 2025
Special Issue in
Genes
Pediatric Rare Diseases: Genetics and Diagnosis
Guest Editor: Antonio F. Martínez‐MonsenyDeadline: 15 September 2025
Special Issue in
Genes
Psychiatric Pharmacogenomics
Guest Editor: Martina HahnDeadline: 15 September 2025
Special Issue in
Genes
Animal Models, Genetic and Genomic Studies in Cancer and Its Therapy
Guest Editors: Dongyu Jia, Yinu Wang, Yoichiro TamoriDeadline: 15 September 2025
Topical Collections
Topical Collection in
Genes
Tools for Population and Evolutionary Genetics
Collection Editors: David Alvarez-Ponce, Julie M. Allen, Won C. Yim, Marco Fondi
Topical Collection in
Genes
Study on Genotypes and Phenotypes of Pediatric Clinical Rare Diseases
Collection Editors: Livia Garavelli, Stefano Giuseppe Caraffi
Topical Collection in
Genes
Genetics and Genomics of Hereditary Disorders of Connective Tissue
Collection Editors: Nazli B. Mcdonnell, Bert Callewaert, Clair A. Francomano, Philippe Khau-Van-Kien, Yves Dulac
Topical Collection in
Genes
Genetics and Genomics of Rare Disorders
Collection Editors: Stefania Zampatti, Emiliano Giardina







