Detection of Development-Specific MicroRNAs in Rabbit Embryos and Culture Media: A Potential Biomarker Approach for Embryo Quality Assessment
Abstract
1. Introduction
2. Materials and Methods
2.1. miRNA Selection
2.2. Experimental Animals and Animal Care
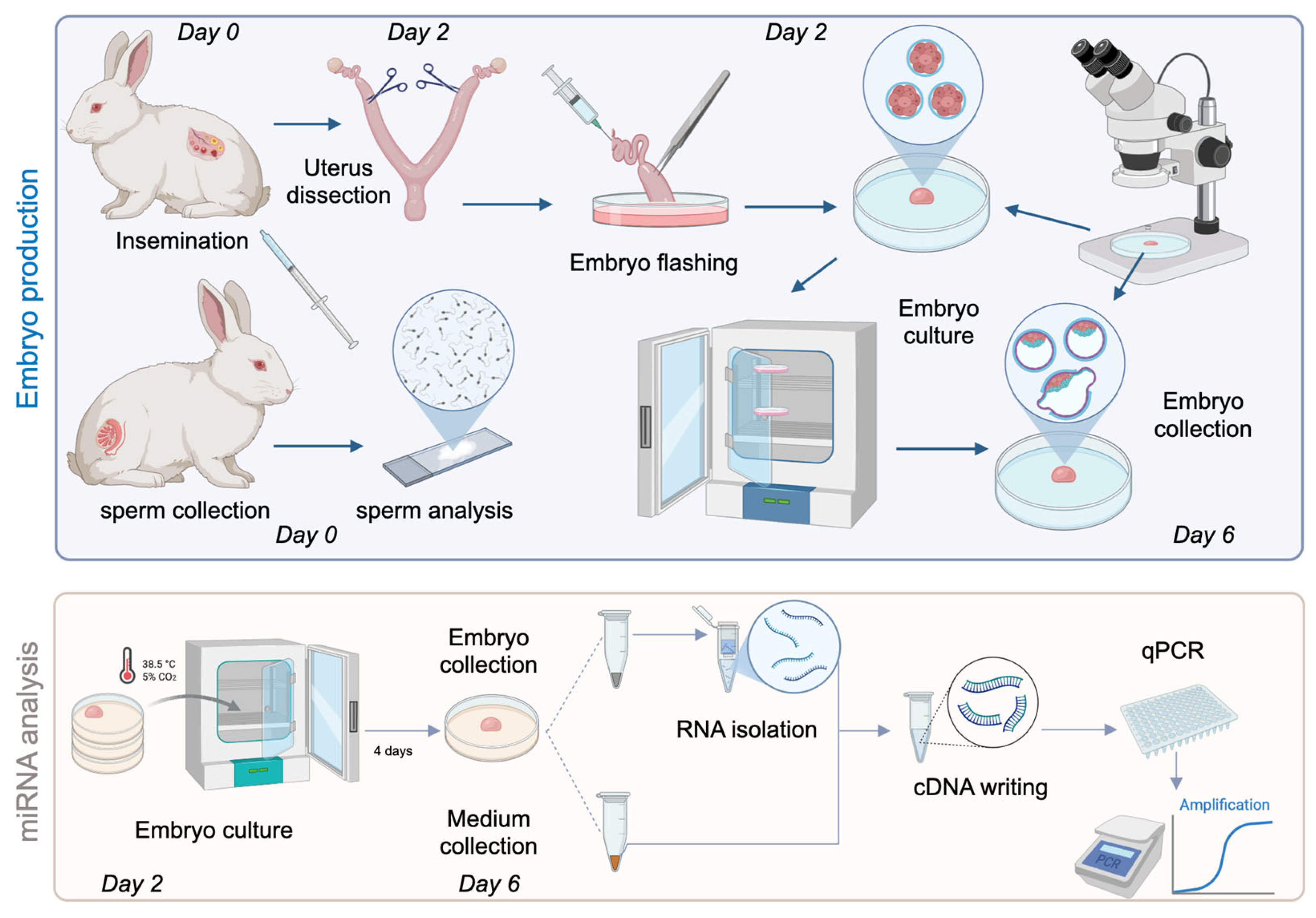
2.3. Rabbit Embryo Collection and Cultivation
2.4. Sample Collection, RNA Isolation, cDNA Writing and qPCR
2.5. Data Analysis—MultiD GenEx
2.6. SOLiD Sequencing
3. Results
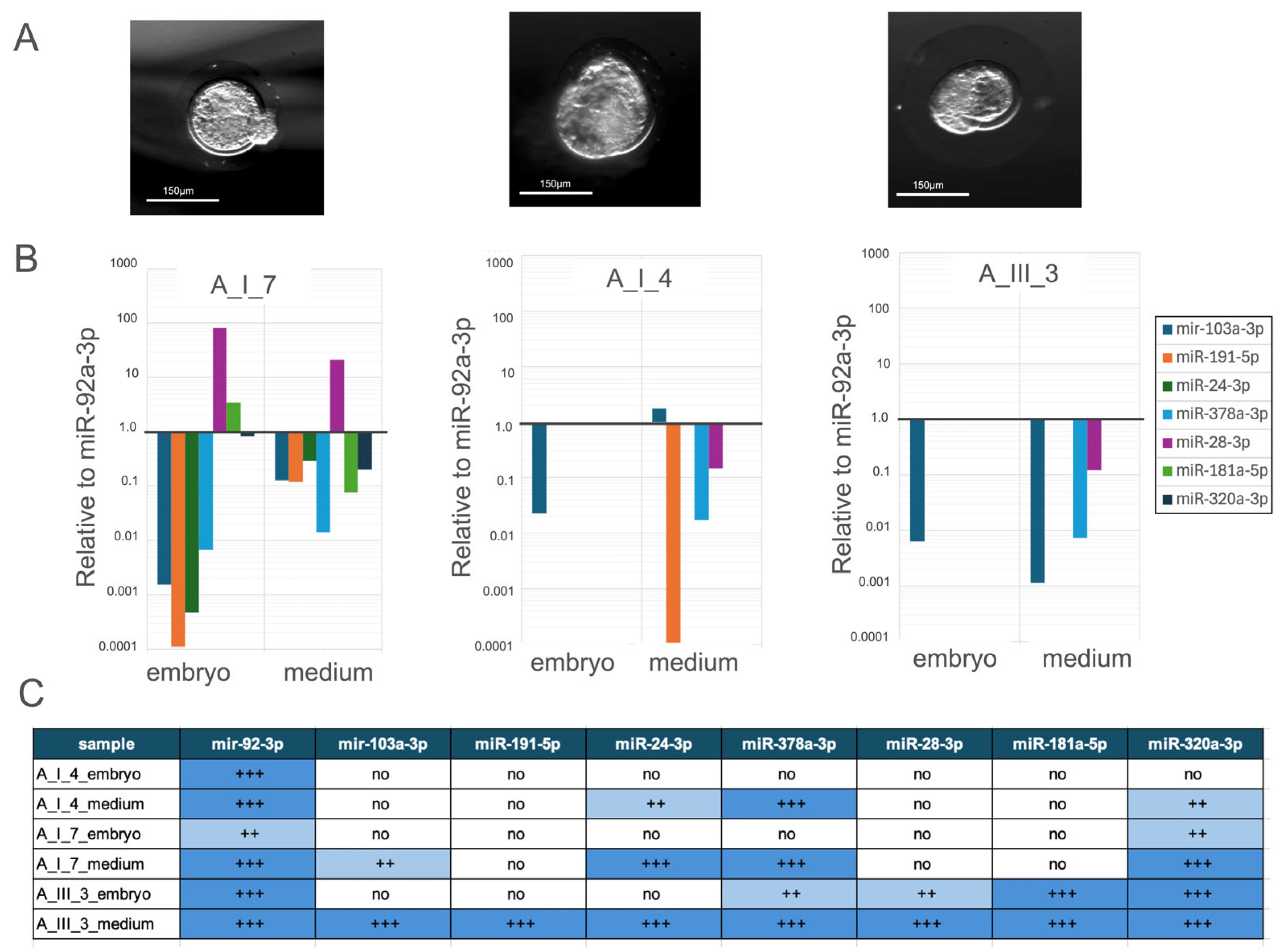
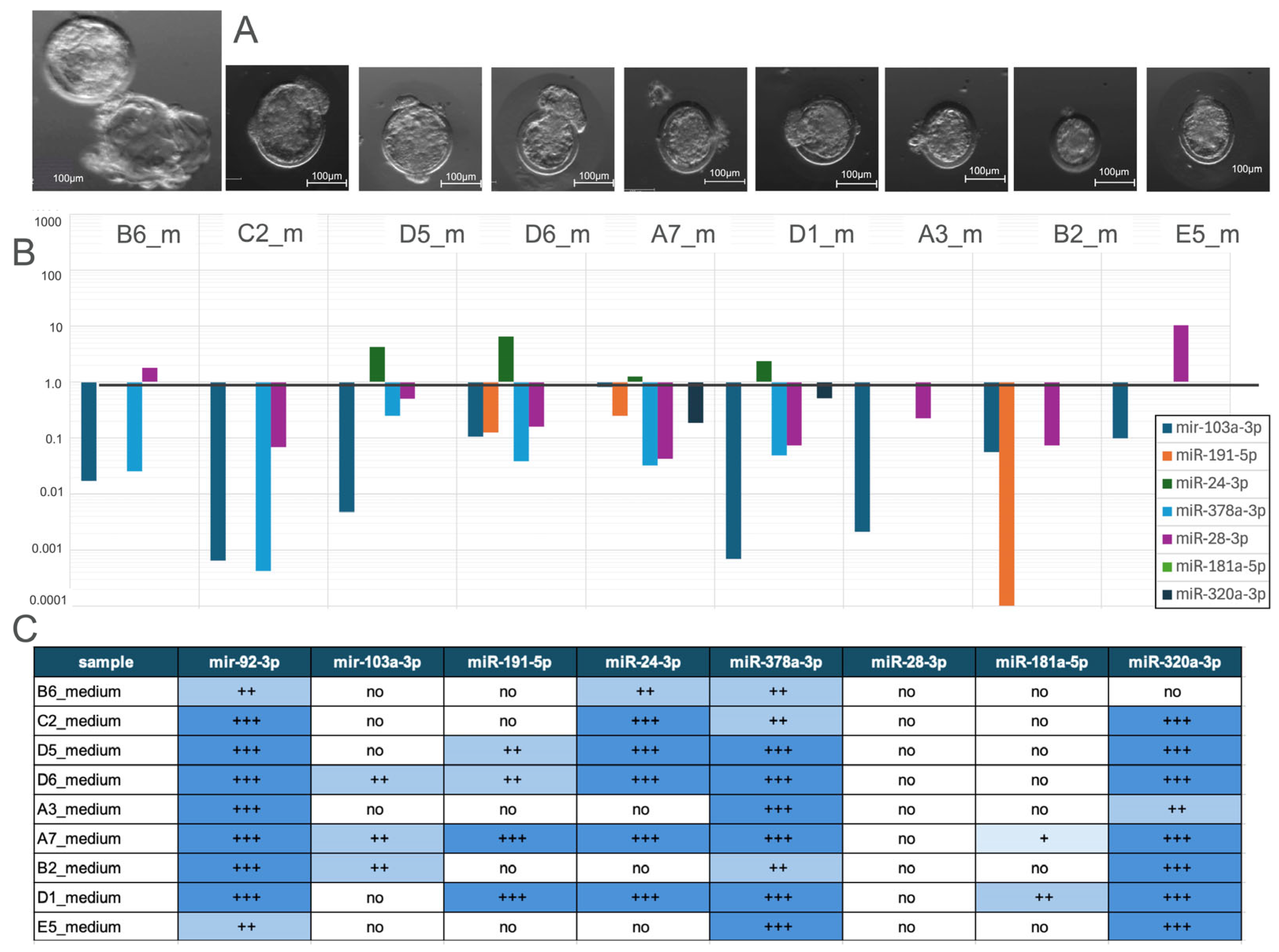
3.1. Sample Collection
3.2. miRNAs Expression Profile
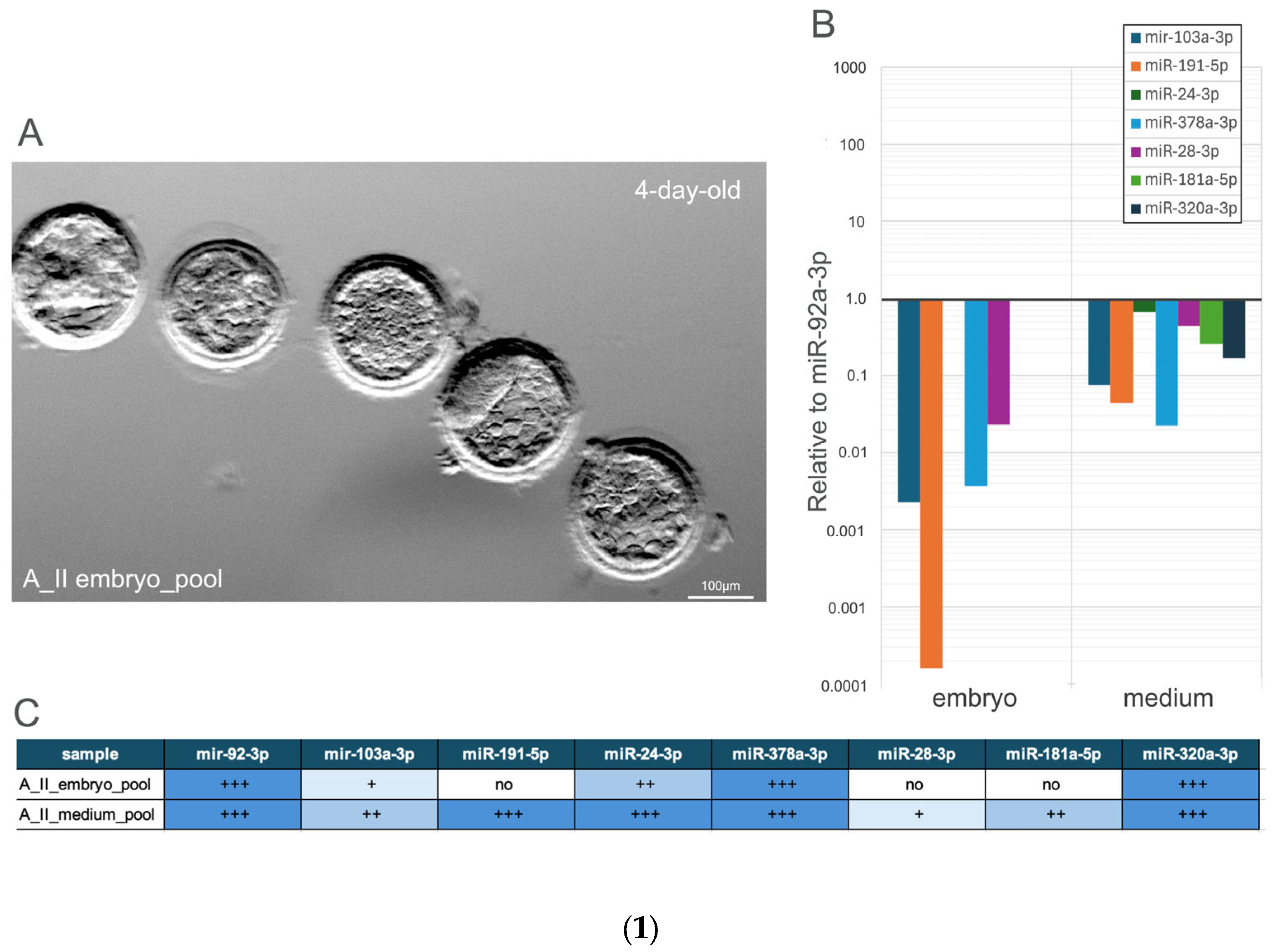
3.2.1. Comparative Analysis of miRNA Expression Profiles in the Blastocysts Pool and Their Culture Medium
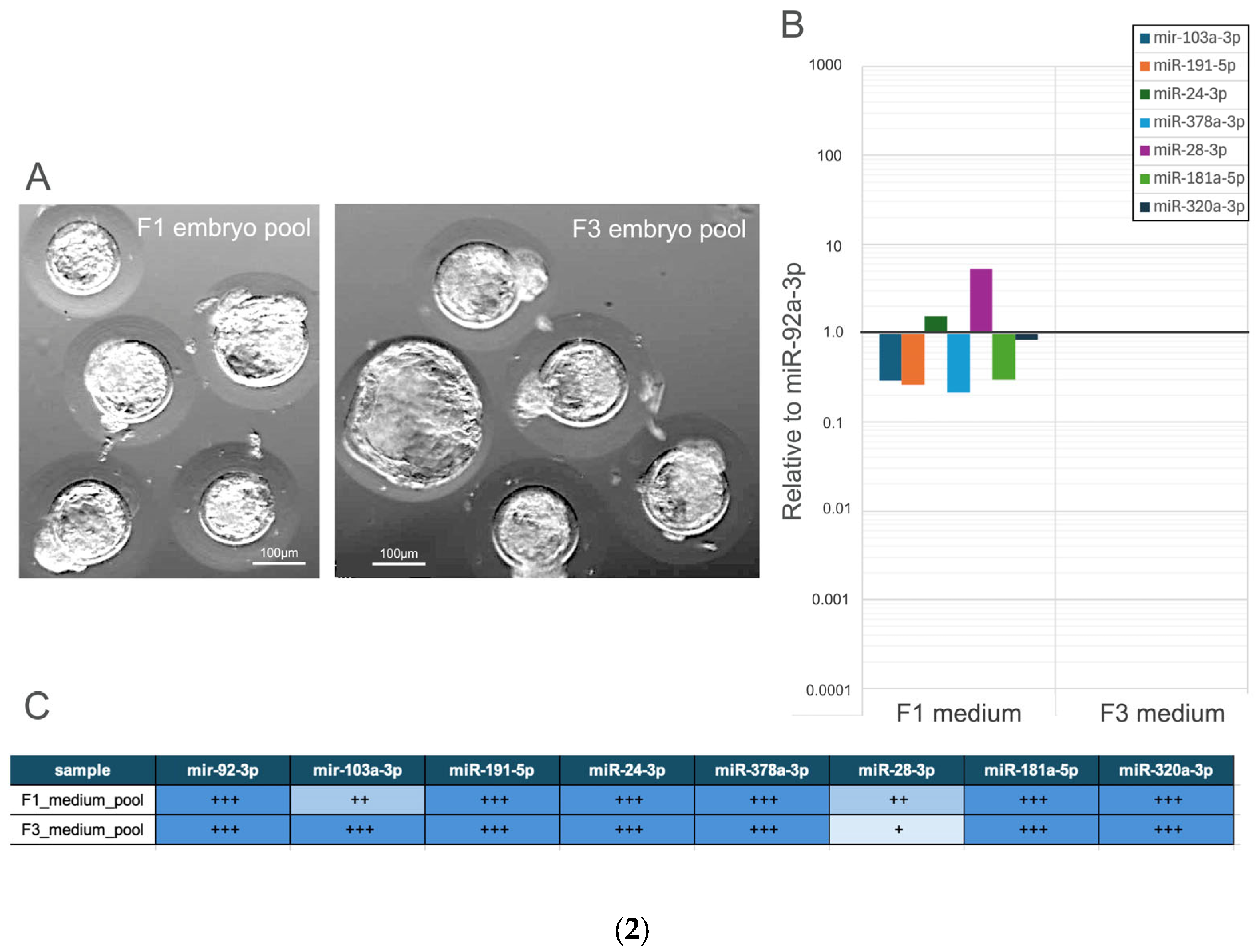
3.2.2. Comparative Analysis of miRNA Expression Profiles in Hatched Blastocysts and Their Culture Medium
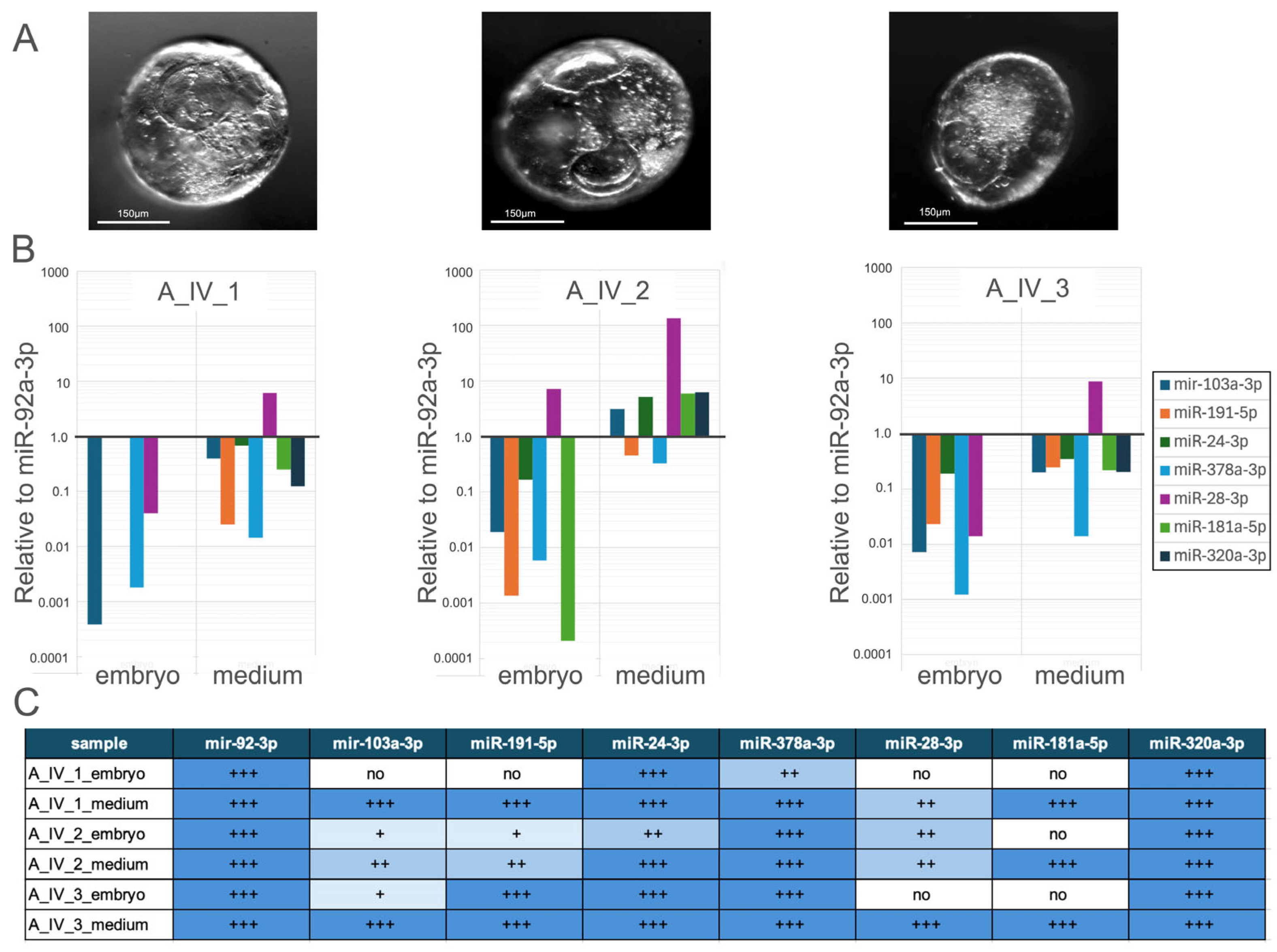
3.2.3. Comparative Analysis of miRNA Expression Profiles in Individually Cultured Blastocysts and Their Culture Medium
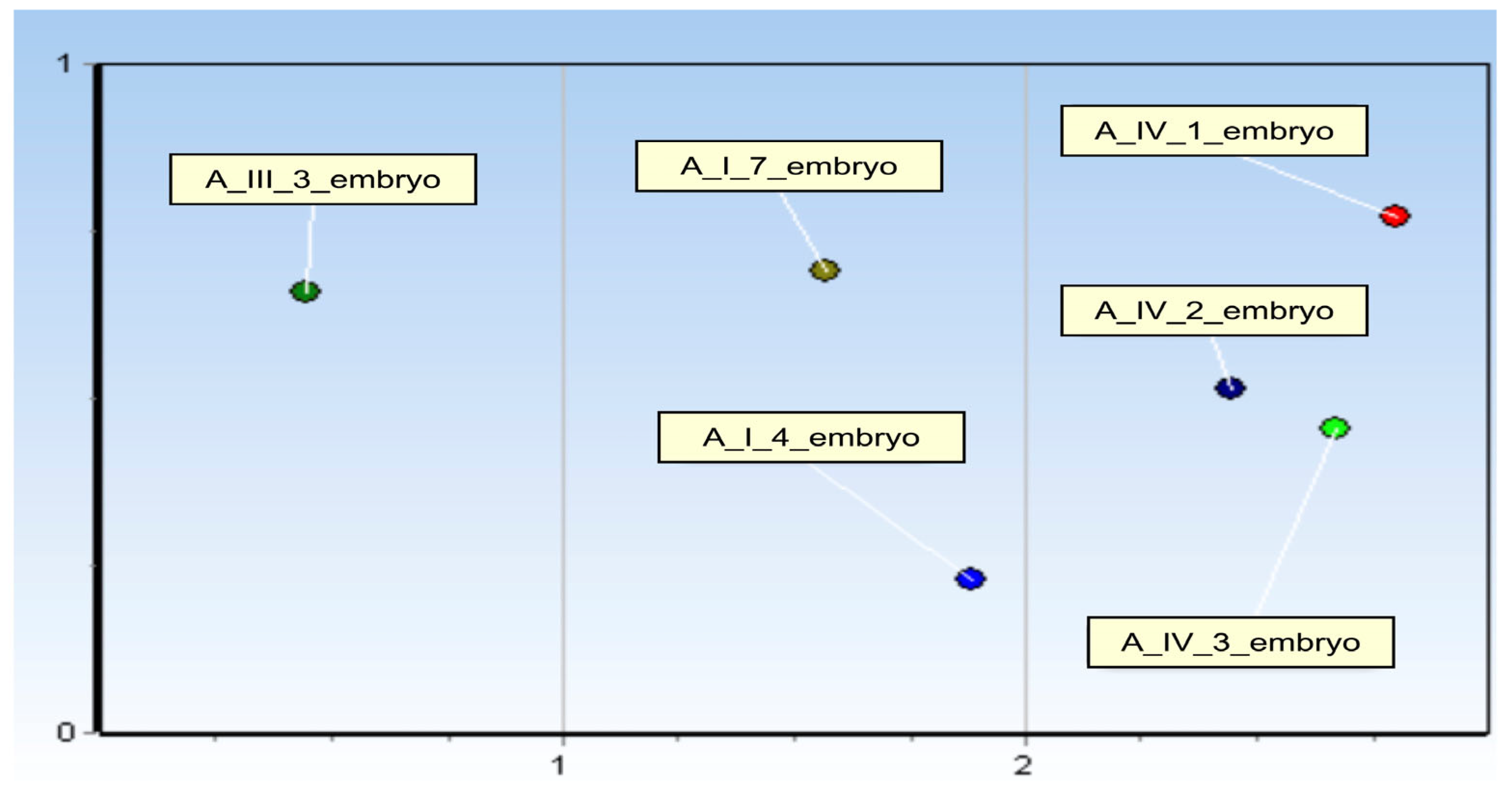
3.2.4. miRNAs Expression Profile—Kohonen’s Self-Organising Map (SOM)
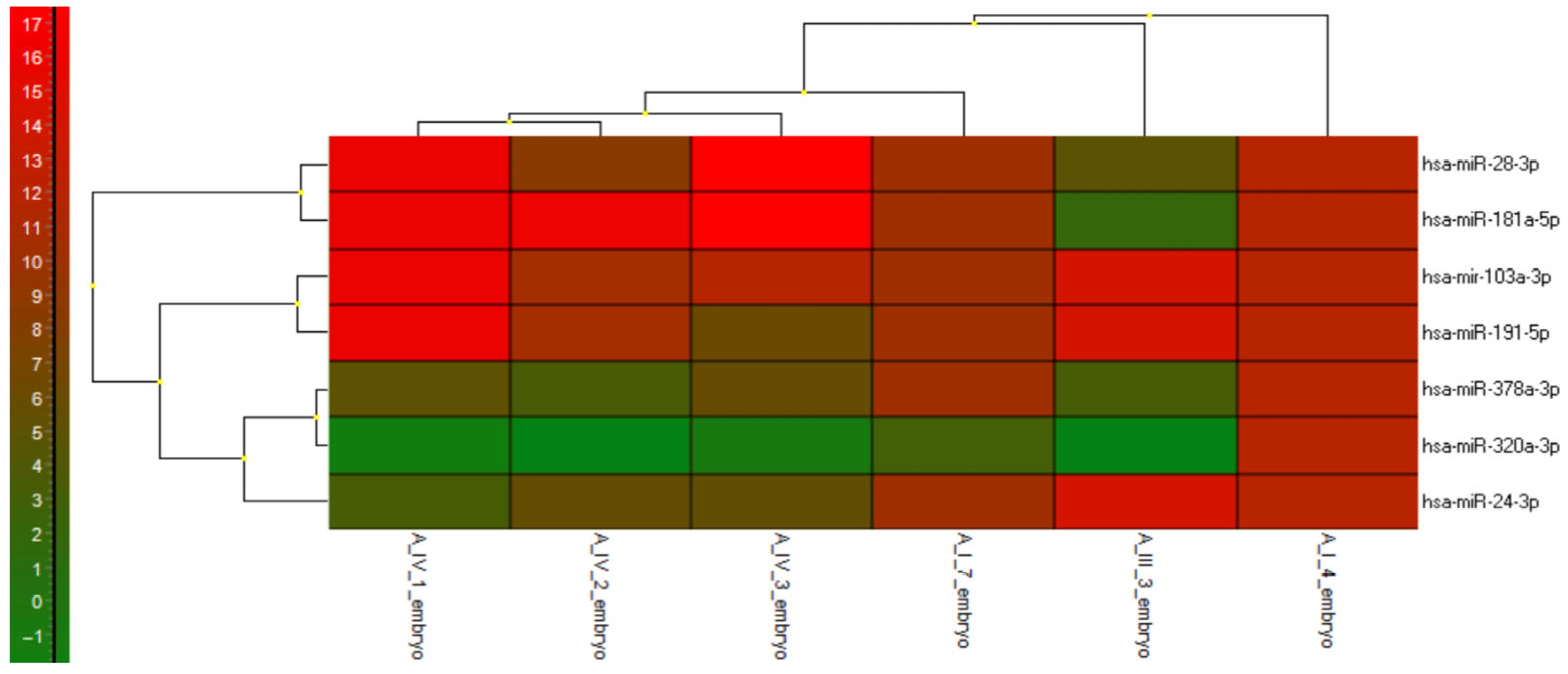
3.2.5. miRNAs Expression Profile—Heatmap
3.2.6. Comparative Analysis of miRNA Expression Profiles in Samples and Their Culture Medium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vidigal, J.A.; Ventura, A. Embryonic stem cell miRNAs and their roles in development and disease. Semin. Cancer Biol. 2012, 22, 428–436. [Google Scholar] [CrossRef]
- Maraghechi, P.; Aponte, M.T.S.; Ecker, A.; Lázár, B.; Tóth, R.; Szabadi, N.T.; Gócza, E. Pluripotency-Associated microRNAs in Early Vertebrate Embryos and Stem Cells. Genes 2023, 14, 1434. [Google Scholar] [CrossRef]
- Lázár, B.; Anand, M.; Tóth, R.; Várkonyi, E.P.; Liptói, K.; Gócza, E. Comparison of the MicroRNA expression profiles of male and female avian primordial germ cell lines. Stem Cells Int. 2018, 2018, 1780679. [Google Scholar] [CrossRef]
- Omes, C.; Conti, A.; Benedetti, L.; Tomasoni, V.; De Marchi, D.; Nappi, R.E.; De Angelis, M.G.C.; Ceccarelli, G. Expression of miRNA from spent pre-implantation embryos culture media. Reprod. Biol. 2024, 24, 100847. [Google Scholar] [CrossRef]
- Mahdipour, M.; van Tol, H.T.; Stout, T.A.; Roelen, B.A. Validating reference microRNAs for normalizing qRT-PCR data in bovine oocytes and preimplantation embryos. BMC Dev. Biol. 2015, 15, 25. [Google Scholar] [CrossRef]
- Benedetti, C.; Pavani, K.C.; Gansemans, Y.; Azari-Dolatabad, N.; Pascottini, O.B.; Peelman, L.; Six, R.; Fan, Y.; Guan, X.; Deserranno, K.; et al. From follicle to blastocyst: microRNA-34c from follicular fluid-derived extracellular vesicles modulates blastocyst quality. J. Anim. Sci. Biotechnol. 2024, 15, 104. [Google Scholar] [CrossRef] [PubMed]
- Pavani, K.C.; Meese, T.; Pascottini, O.B.; Guan, X.; Lin, X.; Peelman, L.; Hamacher, J.; Van Nieuwerburgh, F.; Deforce, D.; Boel, A.; et al. Hatching is modulated by microRNA-378a-3p derived from extracellular vesicles secreted by blastocysts. Proc. Natl. Acad. Sci. USA 2022, 119, e2122708119. [Google Scholar] [CrossRef] [PubMed]
- Turri, F.; Capra, E.; Lazzari, B.; Cremonesi, P.; Stella, A.; Pizzi, F. A Combined Flow Cytometric Semen Analysis and miRNA Profiling as a Tool to Discriminate Between High- and Low-Fertility Bulls. Front. Veter Sci. 2021, 8, 703101. [Google Scholar] [CrossRef] [PubMed]
- Kamijo, S.; Hamatani, T.; Sasaki, H.; Suzuki, H.; Abe, A.; Inoue, O.; Iwai, M.; Ogawa, S.; Odawara, K.; Tanaka, K.; et al. MicroRNAs secreted by human preimplantation embryos and IVF outcome. Reprod. Biol. Endocrinol. 2022, 20, 130. [Google Scholar] [CrossRef]
- Maraghechi, P.; Hiripi, L.; Tóth, G.; Bontovics, B.; Bősze, Z.; Gócza, E. Discovery of pluripotency-associated microRNAs in rabbit preimplantation embryos and embryonic stem-like cells. Reproduction 2013, 145, 421–437. [Google Scholar] [CrossRef][Green Version]
- Mutia, K.; Wiweko, B.; Abinawanto, A.; Dwiranti, A.; Bowolaksono, A. microRNAs as A Biomarker to Predict Embryo Quality Assessment in In Vitro Fertilization. Int. J. Fertil. Steril. 2023, 17, 85–91. [Google Scholar] [CrossRef]
- Khan, H.L.; Bhatti, S.; Abbas, S.; Kaloglu, C.; Isa, A.M.; Younas, H.; Ziders, R.; Khan, Y.L.; Hassan, Z.; Turhan, B.O.; et al. Extracellular microRNAs: Key players to explore the outcomes of in vitro fertilization. Reprod. Biol. Endocrinol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Nadri, P.; Nadri, T.; Gholami, D.; Zahmatkesh, A.; Ghaffari, M.H.; Vargova, K.S.; Savvulidi, F.G.; LaMarre, J. Role of miRNAs in assisted reproductive technology. Gene 2024, 927, 148703. [Google Scholar] [CrossRef]
- SWang, S.; Chen, L.; Zhu, Y.; Jiang, W. Characterization of microRNAs in spent culture medium associated with human embryo quality and development. Ann. Transl. Med. 2021, 9, 1648. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.; Bonetti, T.; de Carvalho, C.; Vigo, F.; Fraietta, R.; Vilella, F.; Simon, C.; Motta, E. miRNA expression in receptive versus post-receptive endometrium in patients undergoing IVF. Fertil. Steril. 2016, 106, e212. [Google Scholar] [CrossRef]
- Zhu, M.; Chu, Y.; Yuan, Q.; Li, J.; Chen, S.; Li, L. Integrated bioinformatics analysis to explore potential therapeutic targets and drugs for small cell carcinoma of the esophagus. Front. Bioinform. 2025, 5, 1495052. [Google Scholar] [CrossRef] [PubMed]
- Sromek, M.; Głogowski, M.; Chechlińska, M.; Kulińczak, M.; Zajdel, M.; Żeber-Lubecka, N.; Bałabas, A.; Szafron, Ł.M.; Kulecka, M.; Siwicki, J.K. Persistent and novel changes in plasma microRNA profiles in patients with non-small cell lung cancer fol-lowing tumour resection. Transl. Lung Cancer Res. 2025, 14, 677–706. [Google Scholar] [CrossRef]
- Hawke, D.C.; Watson, A.J.; Betts, D.H. Extracellular vesicles, microRNA and the preimplantation embryo: Non-invasive clues of embryo well-being. Reprod. Biomed. Online 2021, 42, 39–54. [Google Scholar] [CrossRef]
- Shi, S.; Zhang, L.; Wang, L.; Yuan, H.; Sun, H.; Madaniyati, M.; Cai, C.; Pang, W.; Gao, L.; Chu, G. miR-24-3p promotes proliferation and inhibits apoptosis of porcine granulosa cells by targeting P27. J. Integr. Agric. 2023, 23, 1315–1328. [Google Scholar] [CrossRef]
- Yang, C.; Wang, R.; Hardy, P. The Multifaceted Roles of MicroRNA-181 in Stem Cell Differentiation and Cancer Stem Cell Plasticity. Cells 2025, 14, 132. [Google Scholar] [CrossRef]
- Caporali, A.; Emanueli, C. MicroRNA regulation in angiogenesis. Vasc. Pharmacol. 2011, 55, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Rosenbluth, E.M.; Shelton, D.N.; Sparks, A.E.; Devor, E.; Christenson, L.; Van Voorhis, B.J. MicroRNA expression in the human blastocyst. Fertil. Steril. 2013, 99, 855–861.e3. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.V.; Fedorov, I.S.; Naberezhnev, Y.I.; Tetruashvili, N.K.; Sukhikh, G.T. Key Amniotic Fluid miRNAs as Promising Target Molecules for the Antenatal Prevention of Pulmonary Hypoplasia Associated with Congenital Diaphragmatic Hernia. Int. J. Mol. Sci. 2025, 26, 3872. [Google Scholar] [CrossRef]
- Kotarski, K.; Kot, M.; Skrzypek, K. miR-28: A Tiny Player in Cancer Progression and Other Human Diseases. Biomolecules 2025, 15, 757. [Google Scholar] [CrossRef] [PubMed]
- Pendzialek, S.M.; Knelangen, J.M.; Schindler, M.; Gürke, J.; Grybel, K.J.; Gocza, E.; Fischer, B.; Navarrete Santos, A. Trophoblastic microRNAs are downregulated in a diabetic pregnancy through an inhibition of Drosha. Mol. Cell. Endocrinol. 2019, 480, 167–179. [Google Scholar] [CrossRef]
- Zhou, W.; Dimitriadis, E. Secreted MicroRNA to Predict Embryo Implantation Outcome: From Research to Clinical Diagnostic Application. In Frontiers in Cell and Developmental Biology (Vol. 8). Front. Cell Dev. Biol. 2020, 8, 586510. [Google Scholar] [CrossRef]
| Name of miRNAs | ID Number | miRNA Sequency |
|---|---|---|
| mmu-miR-302b-3p | 481677_mir | UAAGUGCUUCCAUGUUUUAGUAG |
| hsa-miR-92a-3p # | 477827_mir | UAUUGCACUUGUCCCGGCCUGU |
| hsa-miR-378a-3p | 478349_mir | ACUGGACUUGGAGUCAGAAGGC |
| hsa-miR-372-3P | 478071_mir | AAAGUGCUGCGACAUUUGAGCGU |
| hsa-miR-371a-5p | 478851_mir | ACUCAAACUGUGGGGGCACU |
| hsa-miR-320a-3p | 478594_mir | AAAAGCUGGGUUGAGAGGGCGA |
| hsa-miR-302c-3P | 478509_mir | UAAGUGCUUCCAUGUUUCAGUGG |
| hsa-miR-28-3p | 477999_mir | CACUAGAUUGUGAGCUCCUGGA |
| hsa-miR-24-3p | 47932_mir | UGGCUCAGUUCAGCAGGAACAG |
| hsa-miR-191-5p | 477952_mir | CAACGGAAUCCCAAAAGCAGCUG |
| hsa-miR-181a-5p | 477857_mir | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-103a-3p | 478253_mir | AGCAGCAUUGUACAGGGCUAUGA |
| Components | Catalogue Numbers | 96-Well Plate/Well | Final Concentration/Well |
|---|---|---|---|
| TaqMan™ Fast Advanced Master Mix for qPCR (2×) | Applied Biosystems™/4444556 | 10 μL | 1× |
| TaqMan Advanced miRNA Assay/miRNA probe (20×) | Applied Biosystems™/A25576 | 1 μL | 1× |
| cDNA template | Applied Biosystems™/TaqMan™ Advanced miRNA cDNA Synthesis Kit/A28007 | 5 μL | 0.001–100 ng/well |
| Nuclease-free water | Invitrogen™/AM9938 (Invitrogen, Waltham, CA, USA) | 4 μL | |
| Total | 20 μL | ||
| Name of miRNAs | MEF (p2) | REF (p2) | rabESC (p2) | Rabbit Embryo (dpc 6) | Rabbit Embryo (dpc 7) | Rabbit PGCs (dpc 14) |
|---|---|---|---|---|---|---|
| mmu-miR-302b-3p | 0 | 0 | 1 | 1 | 4 | 0 |
| hsa-miR-92a-3p | 125 | 350 | 95 | 61 | 471 | 1540 |
| hsa-miR-378a-3p | 90 | 48 | 21 | 0 | 13 | 16 |
| hsa-miR-372-3P | - | - | - | - | - | - |
| hsa-miR-371a-5p | 0 | 0 | 0 | 7 | 362 | 0 |
| hsa-miR-320a-3p | 70 | 41 | 29 | 6 | 15 | 34 |
| hsa-miR-302c-3p | 0 | 0 | 0 | 2 | 7 | 0 |
| hsa-miR-28-3p | 3 | 8 | 1 | 0 | 0 | 2 |
| hsa-miR-24-3p | 2410 | 2130 | 471 | 3 | 34 | 87 |
| hsa-miR-191-5p | 462 | 555 | 56 | 7 | 187 | 82 |
| hsa-miR-181a-5p | 248 | 96 | 57 | 1 | 3 | 51 |
| hsa-miR-103a-3p | 675 | 662 | 55 | 7 | 90 | 77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salinas, M.; Szabadi, N.T.; Dévai, G.; Urbán, M.; Tóth, A.; Lázár, B.; Pintér, T.; Nemes, A.; Fancsovits, P.; Bodrogi, L.; et al. Detection of Development-Specific MicroRNAs in Rabbit Embryos and Culture Media: A Potential Biomarker Approach for Embryo Quality Assessment. Genes 2025, 16, 1042. https://doi.org/10.3390/genes16091042
Salinas M, Szabadi NT, Dévai G, Urbán M, Tóth A, Lázár B, Pintér T, Nemes A, Fancsovits P, Bodrogi L, et al. Detection of Development-Specific MicroRNAs in Rabbit Embryos and Culture Media: A Potential Biomarker Approach for Embryo Quality Assessment. Genes. 2025; 16(9):1042. https://doi.org/10.3390/genes16091042
Chicago/Turabian StyleSalinas, María, Nikolett Tokodyné Szabadi, Gréta Dévai, Martin Urbán, Arnold Tóth, Bence Lázár, Timea Pintér, Annamária Nemes, Péter Fancsovits, Lilla Bodrogi, and et al. 2025. "Detection of Development-Specific MicroRNAs in Rabbit Embryos and Culture Media: A Potential Biomarker Approach for Embryo Quality Assessment" Genes 16, no. 9: 1042. https://doi.org/10.3390/genes16091042
APA StyleSalinas, M., Szabadi, N. T., Dévai, G., Urbán, M., Tóth, A., Lázár, B., Pintér, T., Nemes, A., Fancsovits, P., Bodrogi, L., & Gócza, E. (2025). Detection of Development-Specific MicroRNAs in Rabbit Embryos and Culture Media: A Potential Biomarker Approach for Embryo Quality Assessment. Genes, 16(9), 1042. https://doi.org/10.3390/genes16091042






