Destructive and Non-Destructive Methods for aDNA Isolation from Teeth and Their Analysis: A Comparison
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
- No. 1.
- Molar teeth of an NN male aged about 50, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław (N = 6).
- No. 2.
- Molar teeth of an NN female aged about 25, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław (N = 6).
- No. 3.
- Molar teeth of an NN male aged about 30, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław (N = 6).
- No. 4.
- Molar teeth of an NN male aged about 45, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław (N = 6).
- No. 5.
- Teeth from the Museum of Forensic Medicine, stored at RT (room temperature) for about 60 years (N = 4).
- No. 6.
- Molar teeth of an NN male aged about 60, obtained on autopsy at the Forensic Medicine Department in Wrocław (N = 4).
- No. 7.
- Skull after preparation with partly preserved dentition. Stored at room temperature (N = 2).
- No. 8.
- Molar teeth of an NN male (accurate evidence material) (N = 2).
- No. 9.
- Molar teeth of an NN male aged about 50, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław, 20 years stored at RT (N = 6).
- No. 10.
- Molar teeth of an NN female aged about 25, obtained on autopsy of a severely putrefied mummified cadaver at the Forensic Medicine Department in Wrocław, 20 years stored at RT (N = 6).
- No. 11.
- Molar teeth of an NN male aged about 30, found in fresh water, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław, 20 years stored at RT (N = 6).
- No. 12.
- Molar teeth of an NN male aged about 45, obtained on autopsy of a severely putrefied cadaver at the Forensic Medicine Department in Wrocław, 20 years stored at RT (N = 6).
- No. 13.
- Molar teeth of an NN male aged about 30, obtained on autopsy of a completely skeletonized cadaver at the Forensic Medicine Department in Wrocław, 20 years stored at RT (N = 4).
- No. 14.
- Molar teeth of an NN male aged about 60, obtained on autopsy at the Forensic Medicine Department in Wrocław, 20 years stored at RT (N = 4).
- No. 15.
- Molar tooth from the Museum of Forensic Medicine, 55 years old, stored at RT (N = 1).
- No. 16.
- Museum skull after chemical whitening and preparation, about 60 years old, male, with partly preserved dentition. Stored at RT (N = 2).
- No. 17.
- Teeth from exhumation of the victim of domestic war, soil grave, 60 years old (Opole, Poland), stored at RT (N = 2).
- No. 18.
- A partially burned tooth from World War II, approximately 65 years old, stored at RT (N = 1).
- No. 19.
- Teeth of an aristocratic Lithuanian family, XVII Century (1562–1620), buried in a common grave in a secret place, after the robbery and devastation of the Grave Chapel by soldiers about 30 years before the last burial (Dubingiai, Lithuania) (N = 13).
- No. 20.
- Molar tooth from a soil grave, XV Century (Wroclaw, Poland), stored at RT (N = 1).
- No. 21.
- Tooth from the Catholic Church Relic, 13th Century (Kraków, Poland), stored at RT (N = 1).
- No. 22.
- Teeth from archeological excavations at the site of Milicz churchyard (XI ng/μL Century). Since the 60s, the material has been stored at room temperature at the Bone Collection of the Museum of Human History, Anthropology Institute, Polish Academy of Sciences in Wrocław (N = 5).
- No. 23.
- Molar teeth, X Century (Ryczyn, Poland), stored at RT (N = 4).
- No. 24.
- Molar tooth of the migration period, 5th century (Kroczycka Cave, Poland), stored at RT (N = 1).
- No. 25.
- Neolithic, molar teeth 4000 BC; Jagodno, Wroclaw, Poland [22] (N = 2).
- No. 26.
- Neanderthal molar tooth, dated approximately 100,000 years (Stajnia Cave, Poland), obtained DNA was repaired and cloned before testing; however, only male AMEL was detected [23] (N = 1).
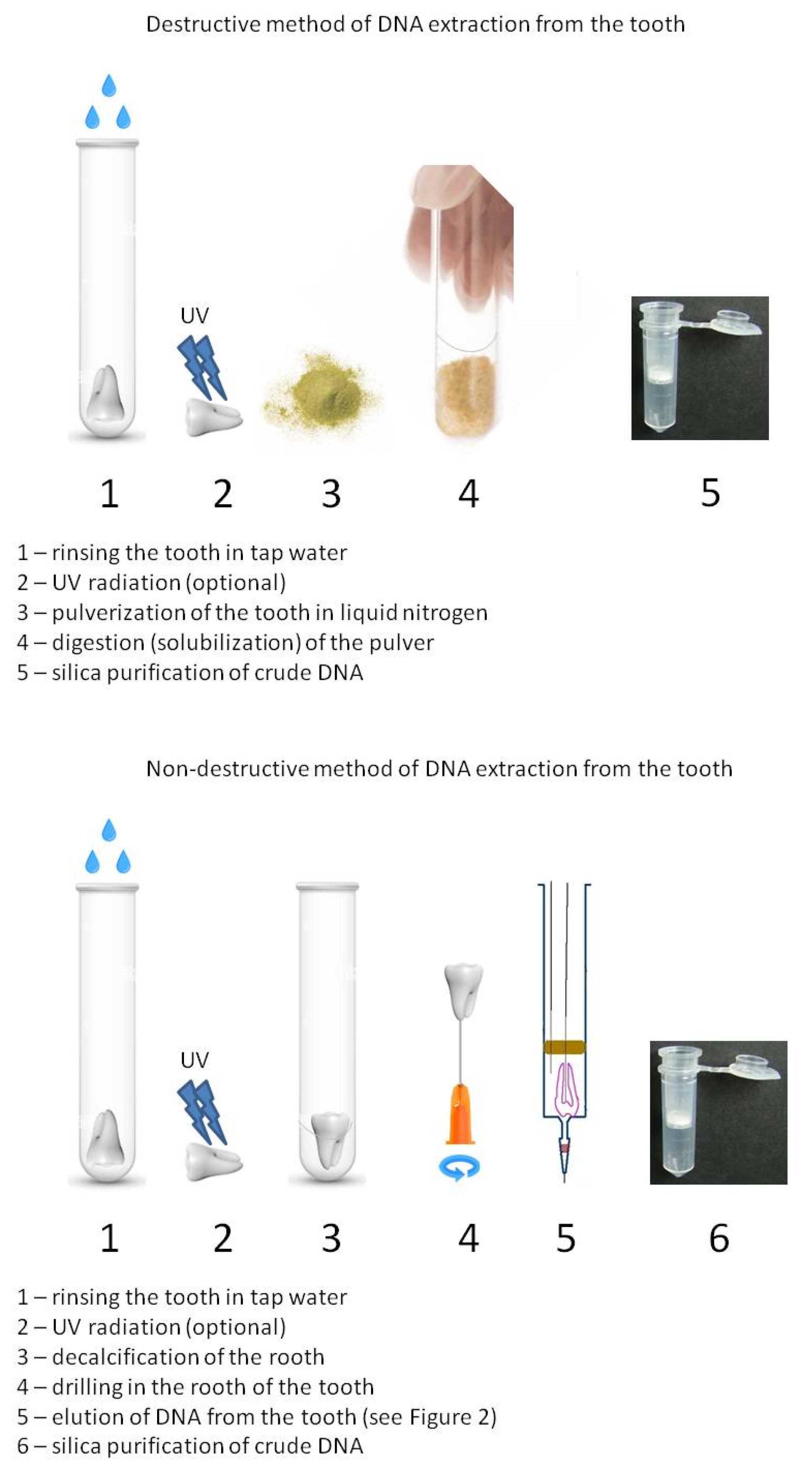
2.2. Destructive Approach
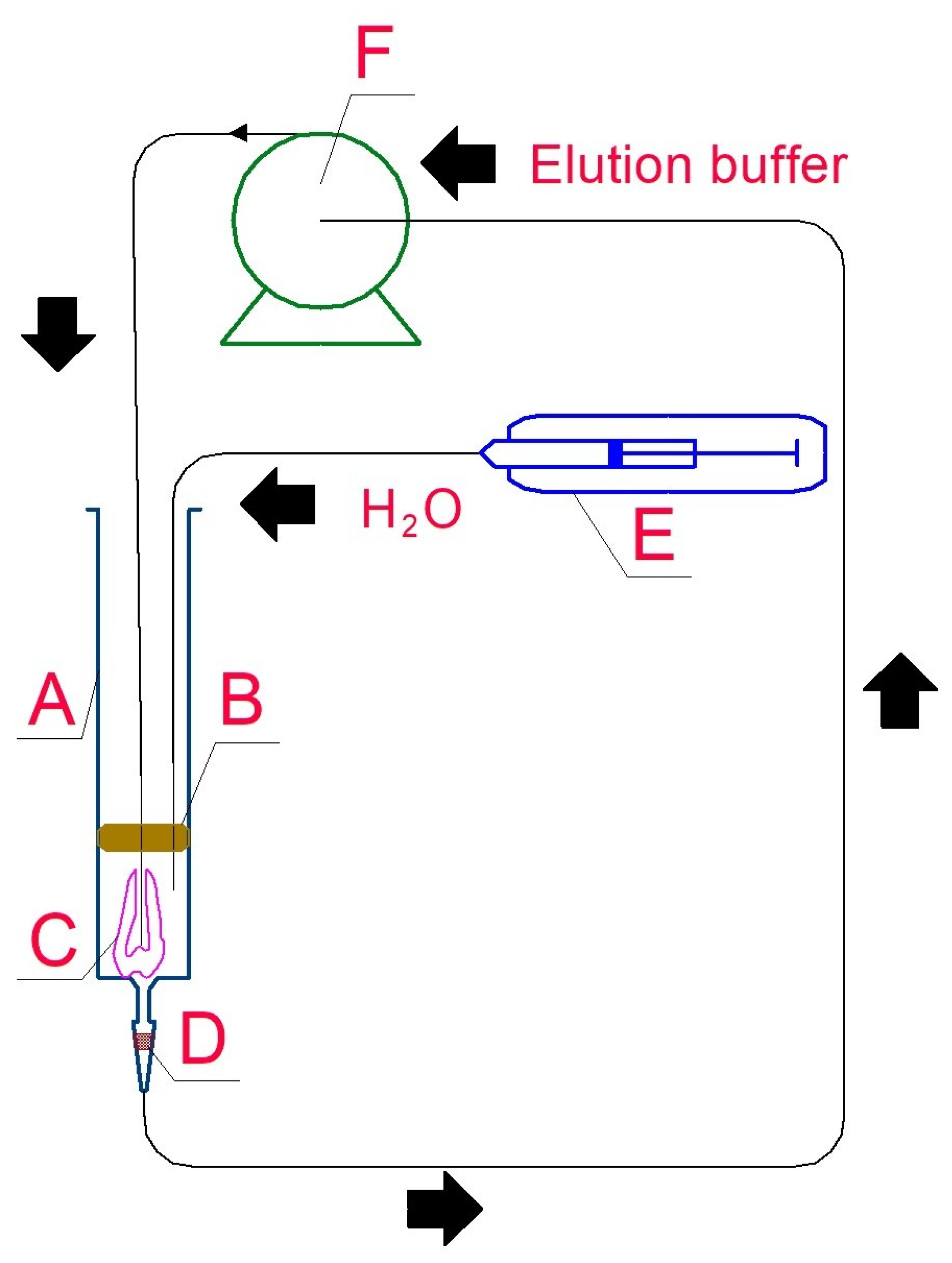
2.3. DNA Extraction Methods
2.4. Purification of Crude DNA
2.5. Quantitative RT PCR
2.6. STR Analysis
2.7. Y-SNP Analysis
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Dun, C.M.P. Non Destructive Procedure for the Isolation of DNA from Plants. The Netherlands Patent No. WO2007093448A1, 23 August 2007. Available online: https://patents.google.com/patent/WO2007093448A1/en (accessed on 7 August 2025).
- Thomsen, P.F.; Elias, S.; Gilbert, T.P.; Haile, J.; Munch, K.; Kuzmina, S.; Froese, D.G.; Sher, A.; Holdaway, R.N.; Willerslev, E. Non-Destructive Sampling of Ancient Insect DNA. PLoS ONE 2009, 4, e5048. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Larralde, A.J.; Suaste-Dzul, A.P.; Gallou, A.; Peña-Carrillo, K.I. DNA recovery from microhymenoptera using six non-destructive methodologies with considerations for subsequent preparation of museum slides. Genome 2017, 60, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Wasko, A.P.; Martins, C.; Oliviera, C.; Forestil, F. Non-destructive genetic sampling in fish. An improved method for DNA extraction from fish fins and scales. Hereditas 2003, 138, 161–165. [Google Scholar] [CrossRef]
- Shepherd, L.D. A non-destructive DNA sampling technique for herbarium specimens. PLoS ONE 2017, 12, e0183555. [Google Scholar] [CrossRef]
- Li, M.; Poonam, A.D.; Cui, Q.; Hsieh, T.; Jagadeesan, S.; Xu, J.; Bruce, W.B.; Vogel, J.T.; Sessions, A.; Cabrera, A.; et al. Non-destructive seed genotyping via microneedle-based DNA extraction. Plant Biotechnol. J. 2025, 23, 2317–2329. [Google Scholar] [CrossRef]
- Bolnick, D.; Bonine, H.; Mata-Míguez, J.; Kemp, B.; Snow, M.; LeBlanc, S. Nondestructive sampling of human skeletal remains yields ancient nuclear and mitochondrial DNA. Am. J. Phys. Anthropol. 2012, 147, 293–300. [Google Scholar] [CrossRef]
- Rayo, E.; Ferrari, G.; Neukamm, J.; Akgül, G.; Breidenstein, A.M.; Cooke, M.; Phillips, C.; Bouwman, A.S.; Rühli, F.J.; Schuenemann, V.J. Non-Destructive Extraction of DNA from Preserved Tissues in Medical Collections. BioTechniques 2022, 72, 60–64. [Google Scholar] [CrossRef]
- Gilbert, M.T.P.; Rudbeck, L.; Willerslev, E.; Hansen, A.J.; Smith, C.; Penkman, K.E.; Prangenberg, K.; Nielsen-Marsh, C.M.; Jans, M.E.; Arthur, P.; et al. Biochemical and physical correlates of DNA contamination in archaeological human bones and teeth excavated at Matera, Italy. J. Arch. Sci. 2005, 32, 785–793. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Pruvost, M.; Schwarz, R.; Correia, V.B.; Champlot, S.; Braguier, S.; Morel, N.; Fernandez-Jalvo, Y.; Grange, T.; Geigl, E.M. Freshly excavated fossil bones are best for amplification of ancient DNA. Proc. Natl. Acad. Sci. USA 2007, 104, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Oota, H.; Saitou, N.; Matsushita, T.; Ueda, S. A genetic study of 2000 year old human remains from Japan using mitochondrial DNA sequences. Am. J. Phys. Anthropol. 1995, 98, 133–145. [Google Scholar] [CrossRef]
- Pinhasi, R.; Fernandes, D.; Sirak, K.; Novak, M.; Connell, S.; Alpaslan-Roodenberg, S.; Gerritsen, F.; Moiseyev, V.; Gromov, A.; Raczky, P.; et al. Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PLoS ONE 2015, 10, e0129102. [Google Scholar] [CrossRef]
- Drancourt, M.; Aboudharam, G.; Croce, O.; Armougom, F.; Robert, C.; Raoult, D. Dental pulp as a source of low-contaminated DNA. Microb. Pathog. 2017, 105, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.C. Ancient DNA recovered by a non destructive method. Anc. Biomol. 2002, 4, 169–172. [Google Scholar] [CrossRef]
- Rohland, N.; Siedel, H.; Hofreiter, M. Nondestructive DNA extraction method for mitochondrial DNA analyses of museum specimens. BioTechniques 2004, 36, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Harney, É.; Cheronet, O.; Fernandes, D.M.; Sirak, K.; Mah, M.; Bernardos, R.; Adamski, N.; Broomandkhoshbacht, N.; Callan, K.; Lawson, A.M.; et al. A minimally destructive protocol for DNA extraction from ancient teeth. Genome Res. 2021, 31, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.; Arooj, A.; Aslam, M.; Cholistani, M.S.; Farhan, M.; Kareem, K.; Ashraf, M.; Pervaiz, M. Methods of Extraction of Genetic Material from Hard Tissues: A Review of the 21st Century Advancements. Forensic Sci. Int. 2025, 367, 112382. [Google Scholar] [CrossRef]
- Hervella, M.; Iñiguez, M.; Izagirre, N.; Anta, A.; De la Rúa, C. Nondestructive Methods for Recovery of Biological Material from Human Teeth for DNA Extraction. J. Forensic Sci. 2014, 60, 136–141. [Google Scholar] [CrossRef]
- Wei, Y.F.; Lin, C.Y.; Yu, Y.J.; Linacre, A.; Lee, J.C. DNA identification from dental pulp and cementum. Forensic Sci. Int. Genet. 2023, 67, 102945. [Google Scholar] [CrossRef] [PubMed]
- Malaver, P.C.; Yunis, J.J. Different dental tissues as source of DNA for human identification in forensic cases. Croat. Med. J. 2003, 44, 306–309. Available online: https://www.cmj.hr/2003/44/3/12808723.pdf (accessed on 28 August 2025).
- Gworys, B.; Rosinczuk-Tonderys, J.; Chrószcz, A.; Janeczek, M.; Dwojak, A.; Bazan, J.; Furmanek, M.; Dobosz, T.; Bonar, M.; Jonkisz, A.; et al. Assessment of late Neolithic pastoralist’s life condition from the Wroclaw-Jagodno site (SW Poland) on the basis of physiological stress markers. J. Archaeol. Sci. 2013, 40, 2621–2630. [Google Scholar] [CrossRef]
- Urbanowski, M.; Socha, P.; Dąbrowski, P.; Nowaczewska, W.; Sadakierska-Chudy, A.; Dobosz, T.; Stefaniak, K.; Nadachowski, A. The first Neanderthal tooth found North of the Carpathian Mountains. Naturwissenschaften 2010, 97, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Hofreiter, M. Ancient DNA extraction from bones and teeth. Nat. Protocols. 2007, 2, 1756–1762. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Cooper, A.; Poinar, H.N. Ancient DNA: Do it right or not at all. Science 2000, 289, 1139–1142. [Google Scholar] [CrossRef]
- Hebsgaard, M.B.; Phillips, M.J.; Willerslev, E. Geologically ancient DNA: Fact or artefact? Trends Microbiol. 2005, 13, 212–220. [Google Scholar] [CrossRef]
- Stoneking, M. Ancient DNA: How do you know when you have it and what can you do with it? Am. J. Hum. Genet. 1995, 57, 1259–1262. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC1801427/pdf/ajhg00038-0006.pdf (accessed on 28 August 2025).
- Willerslev, E.; Cooper, A. Ancient DNA. Proc. R. Soc. B 2005, 272, 3–16. [Google Scholar] [CrossRef]
- Gibbon, V.E.; Penny, C.B.; Strkalj, G.; Ruff, P. Brief communication: Minimally invasive bone sampling method for DNA analysis. Am. J. Phys. Anthropol. 2009, 139, 596–599. [Google Scholar] [CrossRef]
- Kolman, C.J.; Tuross, N. Ancient DNA analysis of human populations. Am. J. Phys. Anthropol. 2000, 111, 5–23. [Google Scholar] [CrossRef]
- Kalmár, T.; Bachrati, C.Z.; Marcsik, A.; Raskó, I. A simple and efficient method for PCR amplifiable DNA extraction from ancient bones. Nucleic Acids Res. 2000, 28, e67. [Google Scholar] [CrossRef]
- Żołędziewska, M.; Gronkiewicz, S.; Dobosz, T. Comparison of various decalcificators in preparation of DNA from human rib bones. Anthropol. Rev. 2002, 65, 75–80. [Google Scholar] [CrossRef]
- Lessig, R.; Edelmann, J.; Thiele, K.; Kozhemyako, V.; Jonkisz, A.; Dobosz, T. Results of Y-SNP typing in three different populations. Forensic Sci. Int. Genet. Suppl. Ser. 2008, 1, 219–221. [Google Scholar] [CrossRef]
- Gomes, C.; Palomo-Díez, S.; Roig, J.; López-Parra, A.M.; Baeza-Richer, C.; Esparza-Arroyo, A.; Gibaja, J.; Arroyo-Pardo, A. Nondestructive extraction DNA method from bones or teeth, true or false? Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e279–e282. [Google Scholar] [CrossRef]
- Allentoft, M.E.; Collins, M.; Harker, D.; Haile, J.; Oskam, C.L.; Hale, M.L.; Campos, P.F.; Samaniego, J.A.; Gilbert, M.T.P.; Willerslev, E.; et al. The half-life of DBA in bone: Measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B 2015, 279, 4724–4733. [Google Scholar] [CrossRef]
- Hofreiter, M.; Serre, D.; Poinar, H.N.; Kuch, M.; Pääbo, S. Ancient DNA. Nat. Rev. Genet. 2001, 2, 353–359. [Google Scholar] [CrossRef]
- Hagelberg, E.; Clegg, J.B. Isolation and characterization of DNA from archaeological bone. Proc. R. Soc. Lond. B 1991, 244, 45–50. [Google Scholar] [CrossRef]
- Cooper, A. Reply to Stoneking: Ancient DNA- How do you really know when you have it? Am. J. Hum. Genet. 1997, 60, 1001–1002. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC1712466/pdf/ajhg00004-0256.pdf (accessed on 28 August 2025).
- Hammer, U.; Bulnheim, U.; Wegener, R. On DNA typing of hard tissues. In Advances in Forensic Haemogenetics; Rittner, C., Schneider, P.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1992; Volume 4. [Google Scholar] [CrossRef]
- Hänni, C.; Laudet, V.; Stehelin, D.; Taberlet, P. Tracking the origins of the cave bear (Ursus spelaeus) by mitochondrial DNA sequencing. Proc. Natl. Acad. Sci. USA 1994, 91, 12336–12340. [Google Scholar] [CrossRef]
- Essel, E.; Zavala, E.I.; Schulz-Kornas, E.; Kozlikin, M.B.; Fewlass, H.; Vernot, B.; Shunkov, M.V.; Derevianko, A.P.; Douka, K.; Barnes, I.; et al. Ancient human DNA recovered from a Palaeolithic pendant. Nature 2023, 618, 328–332. [Google Scholar] [CrossRef]
- Sanz, P.; Prieto, V.; Andres, M.I. PCR Genotyping in Dental Pulp from Old Human Skeletal Remains and Fire Victims. In Advances in Forensic Haemogenetics: Proceedings of the 16th Congress of the International Society for Forensic Haemogenetics (Internationale Gesellschaft für Forensische Hämogenetik e.V.), Santiago de Compostela, Spain, 12–16 September 1995; Carracedo, A., Brinkmann, B., Bär, W., Eds.; Springer: Berlin/Heidelberg, Germany, 1996; Volume 6. [Google Scholar] [CrossRef]
- Essel, E.; Korlević, P.; Meyer, M. A method for the temperature-controlled extraction of DNA from ancient bones. Biotechniques 2021, 71, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Zierdt, H.; Hummel, S.; Herrmann, B. Amplification of human short tandem repeats from medieval teeth and bone samples. Hum. Biol. 1996, 68, 185–199. [Google Scholar] [PubMed]
- Burger, J.; Hummel, S.; Herrmann, B. Palaeogenetics and cultural heritage. Palaeogenetics and cultural heritage. Species determination and STR-genotyping from ancient DNA in art and artefacts. Thermochim. Acta 2000, 365, 141–146. [Google Scholar] [CrossRef]
- Ricaut, F.X.; Keyser-Tracqui, C.; Crubézy, E.; Ludes, B. STR-genotyping from human medieval tooth and bone samples. Forensic Sci. Int. 2005, 151, 31–35. [Google Scholar] [CrossRef]
- Dixon, L.; Dobbins, A.; Pulker, H.; Butler, J.; Vallone, P.; Coble, M.D.; Parson, W.; Berger, B.; Grubwieser, P.; Mogensen, H.; et al. Analysis of artificially degraded DNA using STRs and SNPs—Results of a collaborative European (EDNAP) exercise. Forensic Sci. Int. 2006, 164, 33–44. [Google Scholar] [CrossRef]
- Jankauskas, R. Forensic anthropology and mortuary archaeology in Lithuania. Anthr. Anz. 2009, 67, 391–405. [Google Scholar] [CrossRef]
- Tin, M.M.; Economo, E.P.; Mikheyev, A.S. Sequencing degraded DNA from non-destructively sampled museum specimens for RAD-tagging and low-coverage shotgun phylogenetics. PLoS ONE 2014, 9, e96793. [Google Scholar] [CrossRef] [PubMed]
- Juras, A.; Chyleński, M.; Krenz-Niedbała, M.; Malmström, H.; Ehler, E.; Pospieszny, Ł.; Łukasik, S.; Bednarczyk, J.; Piontek, J.; Jakobsson, M.; et al. Investigating kinship of Neolithic post-LBK human remains from Krusza Zamkowa, Poland using ancient DNA. Forensic Sci. Int. Genet. 2017, 26, 30–39. [Google Scholar] [CrossRef] [PubMed]


| No. of Sample (See List in Paragraph Section 2.1.) | Temp. of Elution | Method of DNA Purification from Crude Extract | DNA Yield (μg) | Concentration of Prepared DNA (ng/μL) | DNA Average Concentration, (ng/μL) ± SD |
|---|---|---|---|---|---|
| No. 1 | 37 °C | silica suspension | 0.0015 | 0.03 | 0.04 ± 0.02 |
| 56 °C | silica suspension | 0.004 | 0.08 | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.002 | 0.04 | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.0025 | 0.05 | ||
| 37 °C | phenol–chloroform | 0.001 | 0.02 | ||
| 56 °C | phenol–chloroform | 0.001 | 0.02 | ||
| No. 2 | 37 °C | silica suspension | 0.01 | 0.20 | 0.36 ± 0.12 |
| 56 °C | silica suspension | 0.027 | 0.53 | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.02 | 0.40 | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.02 | 0.40 | ||
| 37 °C | phenol–chloroform | 0.013 | 0.26 | ||
| 56 °C | phenol–chloroform | 0.018 | 0.35 | ||
| No. 3 | 37 °C | silica suspension | 0.0025 | 0.05 | 0.03 ± 0.03 |
| 56 °C | silica suspension | 0.003 | 0.06 | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.001 | 0.02 | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.0015 | 0.03 | ||
| 37 °C | phenol–chloroform | 0 | 0.0 | ||
| 56 °C | phenol–chloroform | 0 | 0.0 | ||
| No. 4 | 37 °C | silica suspension | 0.005 | 0.10 | 0.09 ± 0.02 |
| 56 °C | silica suspension | 0.006 | 0.12 | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.0045 | 0.09 | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.005 | 0.10 | ||
| 37 °C | phenol–chloroform | 0.003 | 0.06 | ||
| 56 °C | phenol–chloroform | 0.003 | 0.06 | ||
| No. 5 | 56 °C | 0.5 M EDTANa2 pH 8.0 + 0.5% Tween®20 + 50 mM DTT + proteinase K | 0.076 | 1.52 | 1.11 ± 0.29 |
| 0.5 M EDTANa2 pH 8.0 + 0.5% Tween®20 + proteinase K | 0.052 | 1.03 | |||
| 0.5 M EDTANa2 pH 8.0 + proteinase K | 0.044 | 0.87 | |||
| 0.5 M EDTANa2 pH 8.0 + 20% SDS + 1 M Tris-HCl. 5 M NaCl + proteinase K | 0.05 | 1.0 | |||
| No. 6 | 56 °C | 0.5 M EDTANa2 pH 8.0 + 0.5% Tween®20 + 50 mM DTT + proteinase K | 0.017 | 0.33 | 0.21 ± 0.09 |
| 0.5 M EDTANa2 pH 8.0 + 0.5% Tween®20 + proteinase K | 0.011 | 0.22 | |||
| 0.5 M EDTANa2 pH 8.0 + proteinase K | 0.006 | 0.12 | |||
| 0.5 M EDTANa2 pH 8.0 + 20% SDS + 1 M Tris-HCl. 5 M NaCl + proteinase K | 0.009 | 0.17 | |||
| No. 7 | 56 °C | 20 μL proteinase K (0.25 mg/mL) | 0.282 | 5.64 | 6.01 ± 0.52 |
| 20 μL 0.5 M EDTANa2 pH 8.0 + 20 μL proteinase K (0.25 mg/mL) | 0.319 | 6.38 | |||
| No. 8 | 56 °C | 20 μL proteinase K (0.25 mg/mL) | 0.72 | 14.31 | 14.92 ± 0.86 |
| 20 μL 0.5 M EDTANa2 pH 8.0 + 20 μL proteinase K (0.25 mg/mL) | 0.78 | 15.53 |
| No. of Sample (See List in Paragraph Section 2.1.) | Elution Temp. | Purification Method | Total Yield (μg) | aDNA Concentration (ng/μL) | Average aDNA Concentration ± SD | Results (bp) | ||
|---|---|---|---|---|---|---|---|---|
| 100 | 200 | 300 | ||||||
| No. 8 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.716 | 14.31 | 14.92 ± 0.86 | + | + | + |
| 0.777 | 15.53 | + | + | + | ||||
| No. 9 | 37 °C | DNA Extraction Kit FERMENTAS | 0.002 | 0.03 | 0.04 ± 0.02 | + | + | − |
| 56 °C | DNA Extraction Kit FERMENTAS | 0.004 | 0.08 | + | + | + | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.002 | 0.04 | + | + | + | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.003 | 0.05 | + | + | + | ||
| 37 °C | phenol–chloroform | 0.001 | 0.02 | + | + | − | ||
| 56 °C | phenol–chloroform | 0.001 | 0.02 | + | + | − | ||
| No. 10 | 37 °C | DNA Extraction Kit FERMENTAS | 0.010 | 0.20 | 0.31 ± 0.13 | + | + | − |
| 56 °C | DNA Extraction Kit FERMENTAS | 0.027 | 0.53 | + | + | + | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.020 | 0.40 | + | + | − | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.020 | 0.40 | + | + | + | ||
| 37 °C | phenol–chloroform | 0.014 | 0.26 | + | + | − | ||
| 56 °C | phenol–chloroform | 0.018 | 0.35 | + | + | − | ||
| No. 11 | 37 °C | DNA Extraction Kit FERMENTAS | 0.003 | 0.05 | 0.027 ± 0.025 | + | + | − |
| 56 °C | DNA Extraction Kit FERMENTAS | 0.003 | 0.06 | + | + | − | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.001 | 0.02 | + | − | − | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.002 | 0.03 | + | + | − | ||
| 37 °C | phenol–chloroform | 0 | 0 | + | − | − | ||
| 56 °C | phenol–chloroform | 0 | 0 | + | − | − | ||
| No. 12 | 37 °C | DNA Extraction Kit FERMENTAS | 0.005 | 0.10 | 0.09 ± 0.02 | + | + | + |
| 56 °C | DNA Extraction Kit FERMENTAS | 0.006 | 0.12 | + | + | + | ||
| 37 °C | QIAamp DNA Micro Kit (Qiagen) | 0.005 | 0.09 | + | + | + | ||
| 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.005 | 0.10 | + | + | + | ||
| 37 °C | phenol–chloroform | 0.003 | 0.06 | + | + | + | ||
| 56 °C | phenol–chloroform | 0.003 | 0.06 | + | + | + | ||
| No. 13 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.076 | 1.52 | 1.11 ± 0.29 | + | + | + |
| 0.052 | 1.03 | + | + | + | ||||
| 0.044 | 0.87 | + | + | + | ||||
| 0.050 | 1.0 | + | + | + | ||||
| No. 14 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.017 | 0.33 | 0.21 ± 0.09 | + | + | + |
| 0.011 | 0.22 | + | + | + | ||||
| 0.006 | 0.12 | + | + | + | ||||
| 0.009 | 0.17 | + | + | + | ||||
| No. 15 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.092 | 1.83 | - | + | + | + |
| No. 16 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.282 | 5.64 | 6.01 ± 0.52 | + | − | − |
| 0.319 | 6.38 | + | − | − | ||||
| No. 17 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.002 | 0.04 | 0.035 ± 0.01 | + | + | − |
| 0.002 | 0.03 | + | + | − | ||||
| No. 18 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.001 | 0.01 | - | + | + | − |
| No. 19 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.469 | 9.37 | - | + | + | − |
| 0.050 | 1.00 | 0.96 ± 1.00 | + | + | − | |||
| 0.002 | 0.03 | − | − | − | ||||
| 0.004 | 0.08 | − | − | − | ||||
| 0.003 | 0.06 | + | + | − | ||||
| 0.003 | 0.05 | + | − | − | ||||
| 0.041 | 0.81 | + | − | − | ||||
| 0.025 | 0.49 | + | − | − | ||||
| 0.052 | 1.03 | − | − | − | ||||
| 0.163 | 3.25 | − | − | − | ||||
| 0.059 | 1.18 | − | − | − | ||||
| 0.055 | 1.09 | − | − | − | ||||
| 0.122 | 2.44 | − | − | − | ||||
| No. 20 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.081 | 1.62 | - | + | + | − |
| No. 21 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.001 | 0.02 | - | + | + | − |
| No. 22 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.002 | 0.03 | 0.16 ± 0.13 | + | − | − |
| 0.013 | 0.26 | + | − | − | ||||
| 0.018 | 0.35 | + | − | − | ||||
| 0.005 | 0.09 | + | + | − | ||||
| 0.005 | 0.09 | + | + | − | ||||
| No. 23 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0.003 | 0.05 | 0.04 ± 0.02 | + | − | − |
| 0.003 | 0.05 | − | − | − | ||||
| 0.002 | 0.03 | − | − | − | ||||
| 0.001 | 0.01 | − | − | − | ||||
| No. 24 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0 | 0 | - | − | − | − |
| No. 25 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0 | 0 | 0 | + | − | − |
| 0 | 0 | + | − | − | ||||
| No. 26 | 56 °C | QIAamp DNA Micro Kit (Qiagen) | 0 | 0 | 0 | − | − | − |
| Number of Tested Samples (N) | Type of Sample | Results | Rate of Success |
|---|---|---|---|
| 30 | Autopsy teeth (fresh) | 7 × FP, 19 × IR, 4 × NR | 87% |
| 6 | Museal | 4 × FP, 2 × IR, | 100% |
| 27 | Museal tested after 20 years) | 16 × FP, 11 × IR | 100% |
| 13 | Archeology (1562–1620) | 6 × IR, 7 × NR | 54% |
| 2 | Archeology (60 years old) | 2 × IR | 100% |
| 1 | Archeology (World War II) | 1 × IR | 100% |
| 1 | Archeology (XV century) | 1 × IR | 100% |
| 1 | Archeology (XIII century) | 1 × IR | 100% |
| 5 | Archeology (XI century) | 5 × IR | 100% |
| 4 | Archeology (X century) | 2 × IR, 2 × NR | 50% |
| 1 | Archeology (V century) | 1 × NR | 0% |
| 2 | Archeology (4000 B.C.) | 2 × IR | 100% |
| 1 | Archeology (Neanderthal, molar tooth, dated about 50,000–100,000 years old) | 1 × NR | 0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobosz, A.; Jonkisz, A.; Lebioda, A.; Kawecki, J.; Dobosz, T. Destructive and Non-Destructive Methods for aDNA Isolation from Teeth and Their Analysis: A Comparison. Genes 2025, 16, 1059. https://doi.org/10.3390/genes16091059
Dobosz A, Jonkisz A, Lebioda A, Kawecki J, Dobosz T. Destructive and Non-Destructive Methods for aDNA Isolation from Teeth and Their Analysis: A Comparison. Genes. 2025; 16(9):1059. https://doi.org/10.3390/genes16091059
Chicago/Turabian StyleDobosz, Agnieszka, Anna Jonkisz, Arleta Lebioda, Jerzy Kawecki, and Tadeusz Dobosz. 2025. "Destructive and Non-Destructive Methods for aDNA Isolation from Teeth and Their Analysis: A Comparison" Genes 16, no. 9: 1059. https://doi.org/10.3390/genes16091059
APA StyleDobosz, A., Jonkisz, A., Lebioda, A., Kawecki, J., & Dobosz, T. (2025). Destructive and Non-Destructive Methods for aDNA Isolation from Teeth and Their Analysis: A Comparison. Genes, 16(9), 1059. https://doi.org/10.3390/genes16091059






