Feature Papers in Epigenomes
A topical collection in Epigenomes (ISSN 2075-4655).
Viewed by 82187Editors
Interests: SWI/SNF chromatin remodeling enzymes; melanocyte differentiation and regulation of pigementation; epigenetic regulation of the response to ultraviolet radiation; cancer epigenetics
Special Issues, Collections and Topics in MDPI journals
Interests: gene regulation; chromatin; DNA methylation; neurofunction; transcription
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
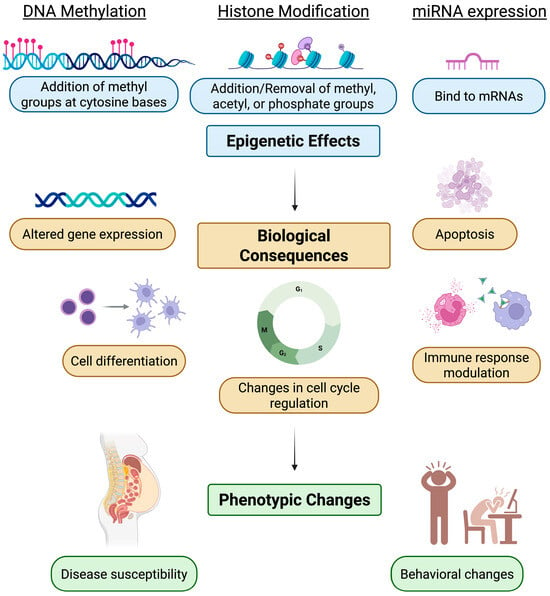
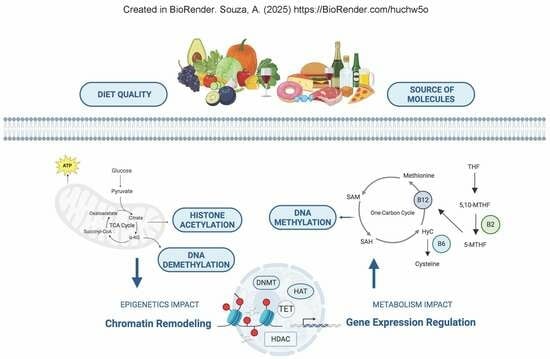
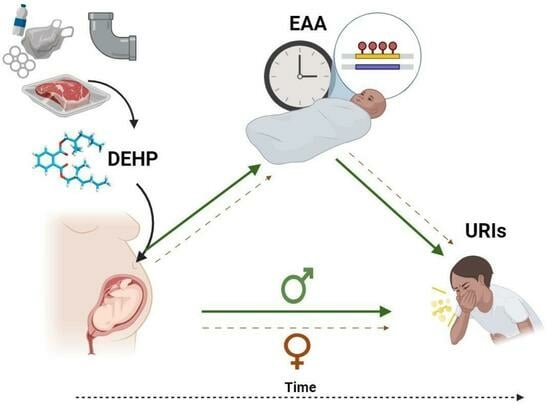
This Topical Collection aims to collect high-quality papers on epigenetics and epigenomics. We encourage researchers and experts from various fields within the journal’s scope to contribute papers that highlight the latest developments in their research field. The potential topics of this Special Issue include, but are not limited to, the following:
- Functional epigenomic studies;
- Genome-wide epigenetic status and the regulation of cells or tissues;
- Chromatin modifications and remodeling in diseases;
- Epigenetics in physical diseases;
- Environmental changes in the epigenetic status of cells or tissues;
- The inheritance or fixation of epigenetic characteristics;
- The description of novel methods to study epigenetic regulation;
- Novel tools, protocols, and technologies for epigenetic studies and therapeutics;
- Long noncoding RNA (LncRNA), microRNA (miR), and chromatin crosstalk.
Dr. Ivana De la Serna
Prof. Dr. Che-Kun James Shen
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Epigenomes is an international peer-reviewed open access quarterly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 1600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- epigenomic
- epigenetic
- chromatin modification
- DNA methylation
- DNA methyltransferases
- cancer epigenetics
- histones