Maternal MicroRNA Profile Changes When LH Is Added to the Ovarian Stimulation Protocol: A Pilot Study
Abstract
:1. Introduction
2. Results
2.1. Profiling of Patients in the Study Groups
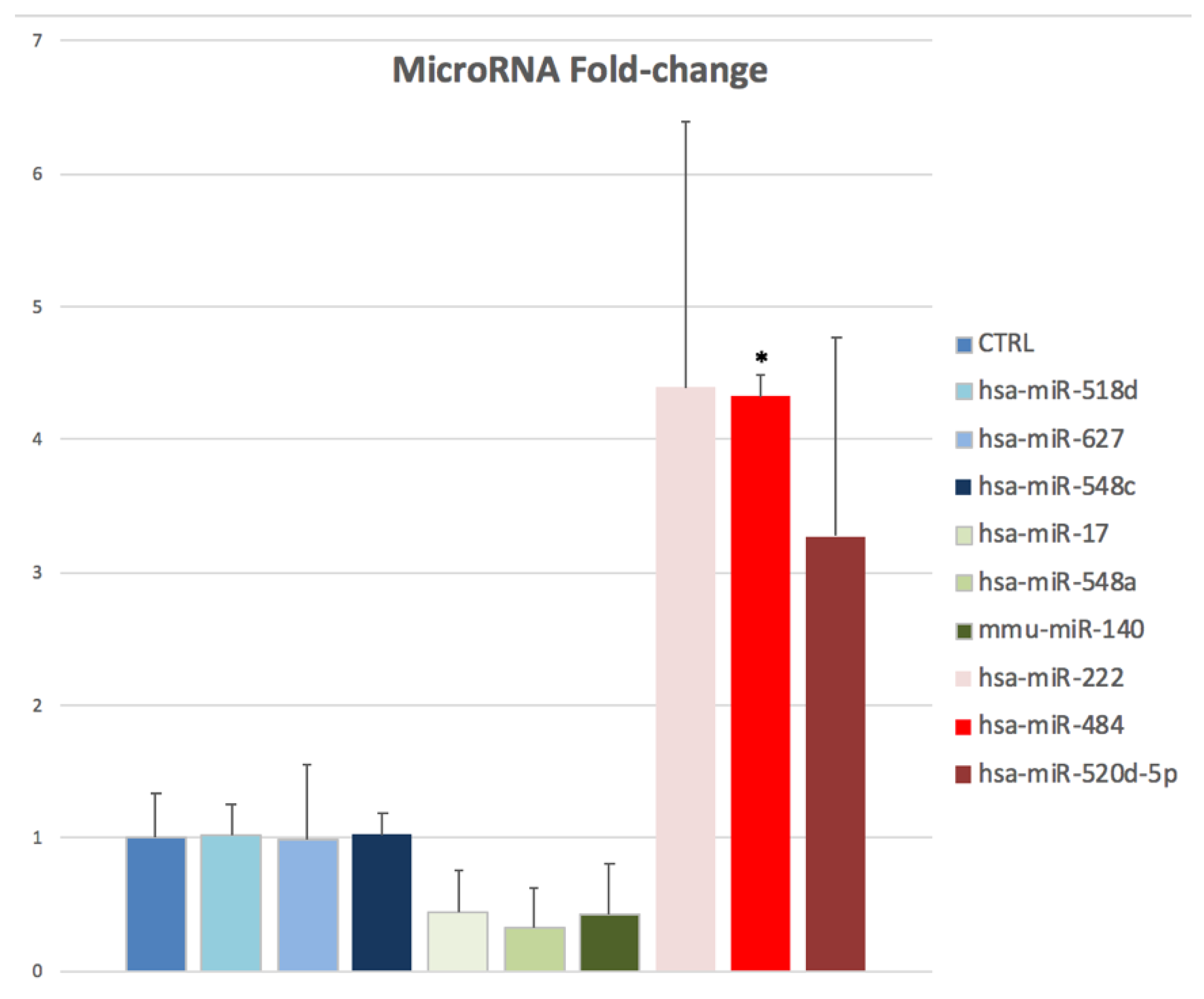
2.2. Expression Patterns of Oocyte miRNAs in the rFSH-rLH Versus the rFSH Treatment Group
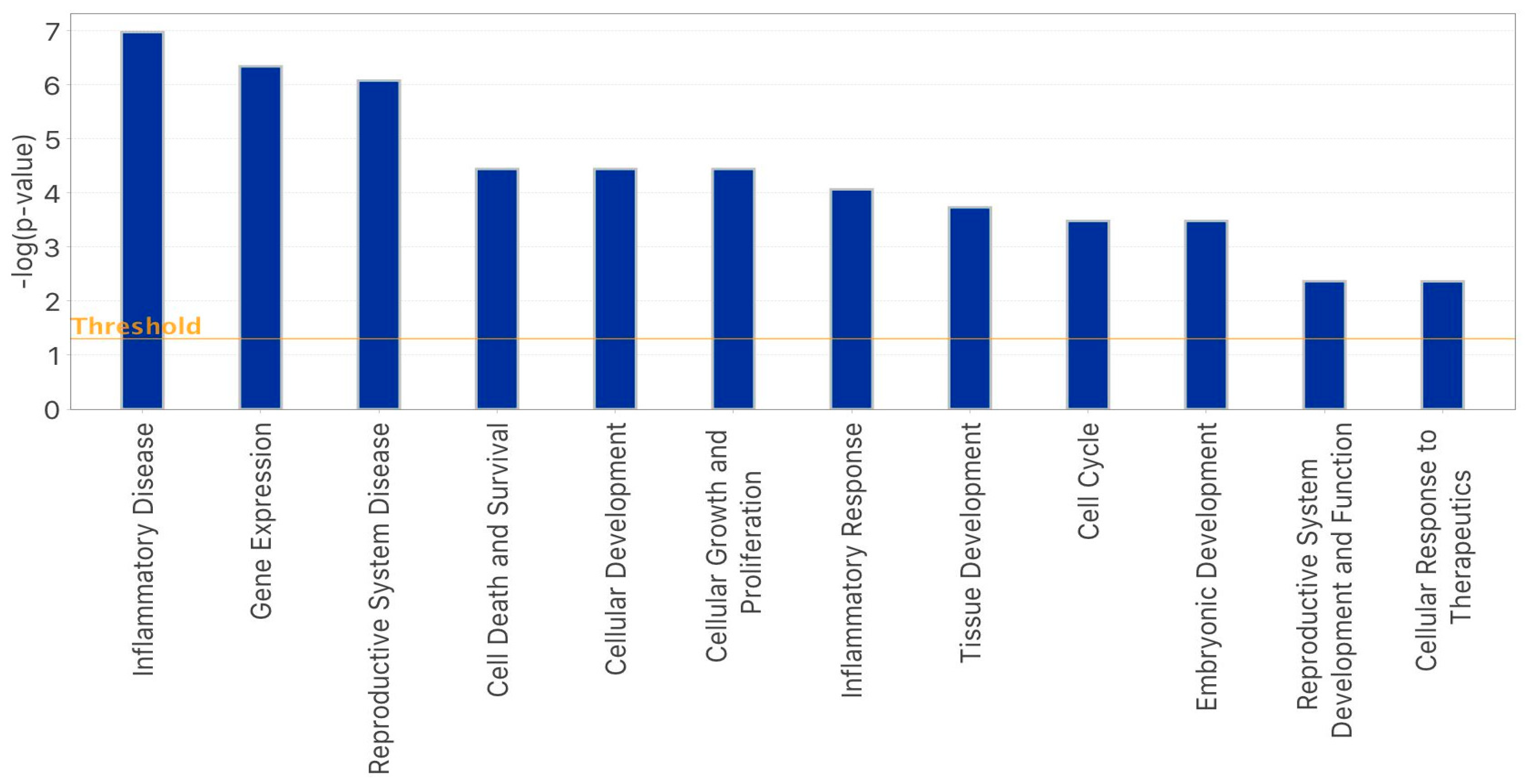
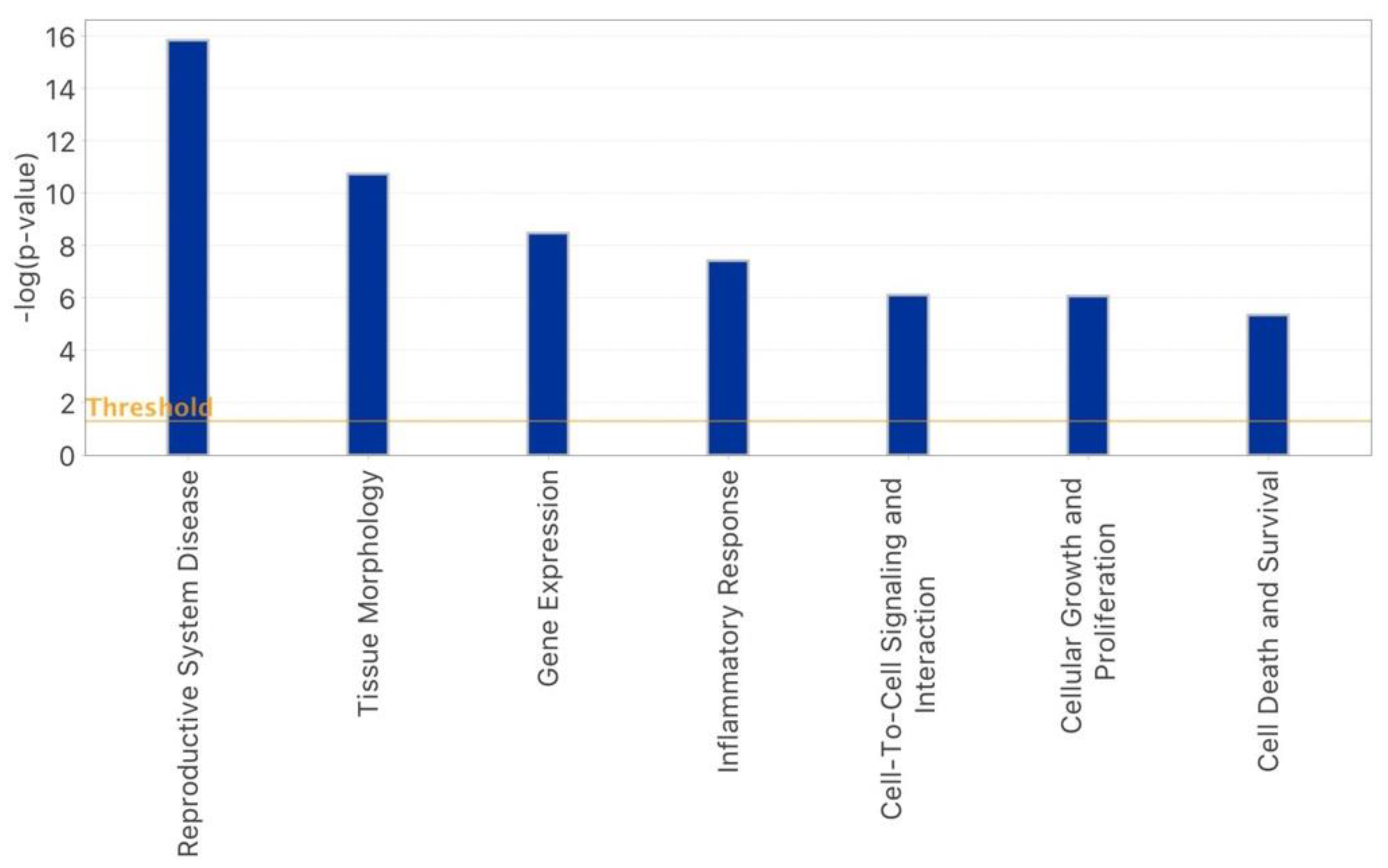
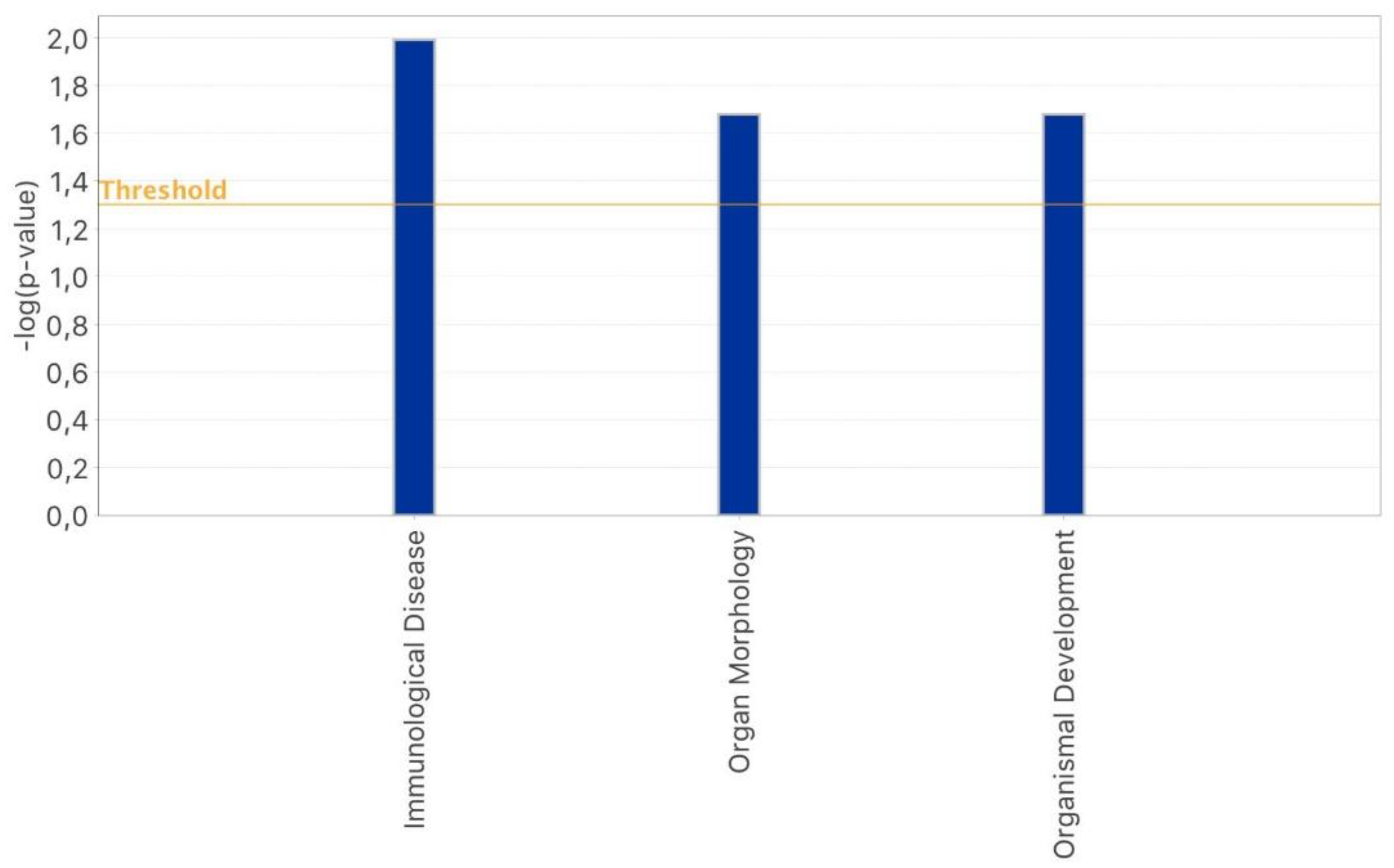
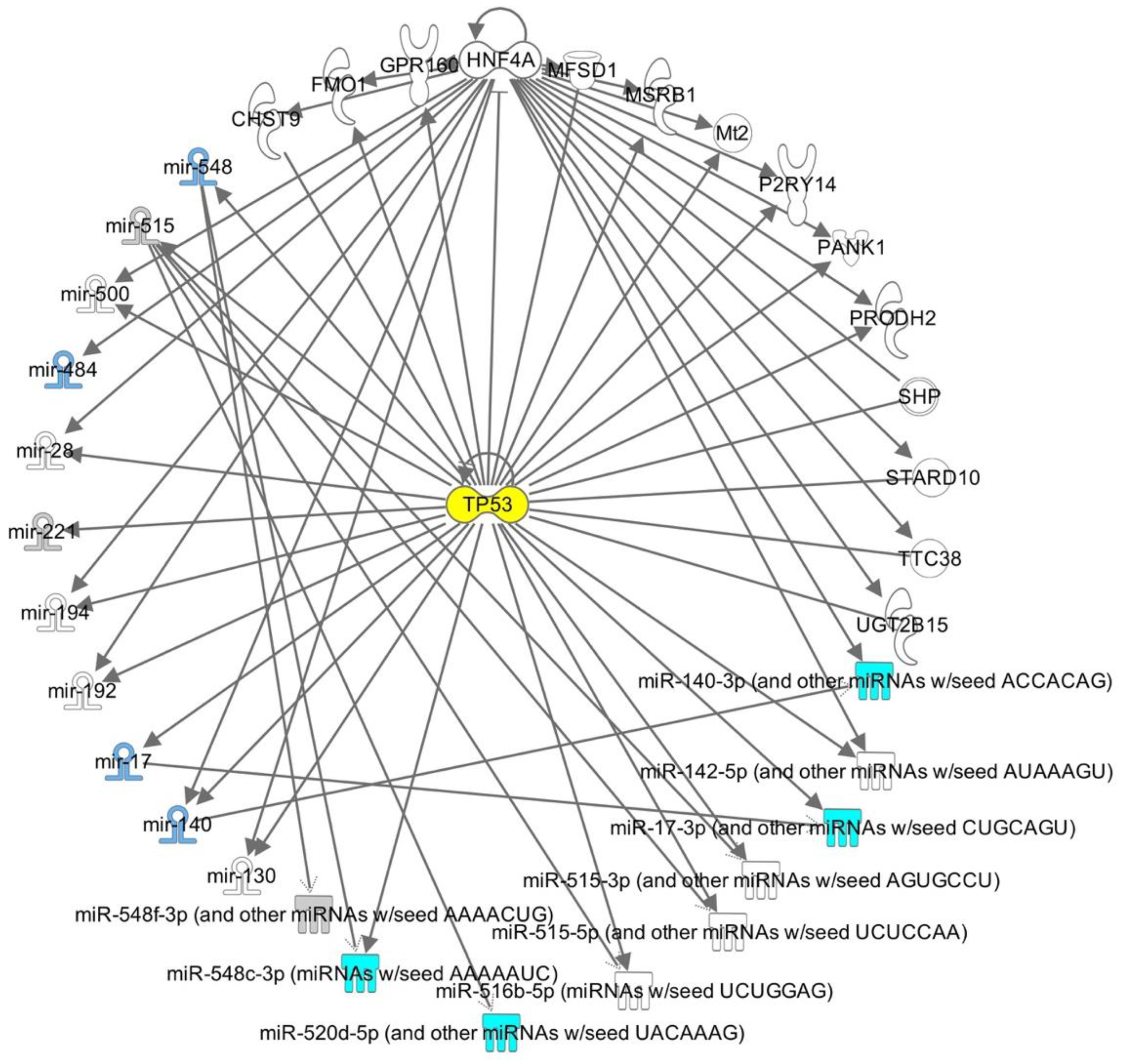
2.3. IPA-Inferred Functional and Network Analysis of Oocyte miRNAs Expressed in rFSH and/or rFSH-rLH Treatment Groups
3. Discussion
4. Materials and Methods
4.1. Patient Population
4.2. Ovarian Stimulation (OS)
4.3. Oocyte Collection
4.4. RNA Extraction from Oocytes
4.5. Characterisation and Expression Profiles of microRNAs in Oocytes of Women Undergoing IVF
4.6. Ingenuity Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jenabi, M.; Khodarahmi, P.; Tafvizi, F.; Bostanabad, S.Z. Evaluation of the Potential of MiR-21 as a Diagnostic Marker for Oocyte Maturity and Embryo Quality in Women Undergoing ICSI. Sci. Rep. 2023, 13, 1440. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.K.S.; Malakar, A.K.; Chakraborty, S. Interplay between MiRNAs and Human Diseases. J. Cell Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Cheung, A.H.-H.; Chan, C.L.-K.; Chan, W.-Y. The Role of MicroRNAs in Ovarian Granulosa Cells in Health and Disease. Front. Endocrinol. 2019, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Revelli, A.; Piane, L.D.; Casano, S.; Molinari, E.; Massobrio, M.; Rinaudo, P. Follicular Fluid Content and Oocyte Quality: From Single Biochemical Markers to Metabolomics. Reprod. Biol. Endocrinol. 2009, 7, 40. [Google Scholar] [CrossRef]
- Moreno, J.M.; Núñez, M.J.; Quiñonero, A.; Martínez, S.; de la Orden, M.; Simón, C.; Pellicer, A.; Díaz-García, C.; Domínguez, F. Follicular Fluid and Mural Granulosa Cells MicroRNA Profiles Vary in in Vitro Fertilization Patients Depending on Their Age and Oocyte Maturation Stage. Fertil. Steril. 2015, 104, 1037–1046.e1. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Blelloch, R. Small RNAs in Early Mammalian Development: From Gametes to Gastrulation. Development 2011, 138, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kaneda, M.; O’Carroll, D.; Hajkova, P.; Barton, S.C.; Sun, Y.A.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. Maternal MicroRNAs Are Essential for Mouse Zygotic Development. Genes Dev. 2007, 21, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Paloviita, P.; Hydén-Granskog, C.; Yohannes, D.A.; Paluoja, P.; Kere, J.; Tapanainen, J.S.; Krjutškov, K.; Tuuri, T.; Võsa, U.; Vuoristo, S. Small RNA Expression and MiRNA Modification Dynamics in Human Oocytes and Early Embryos. Genome Res. 2021, 31, 1474–1485. [Google Scholar] [CrossRef]
- Kataruka, S.; Modrak, M.; Kinterova, V.; Malik, R.; Zeitler, D.M.; Horvat, F.; Kanka, J.; Meister, G.; Svoboda, P. MicroRNA Dilution during Oocyte Growth Disables the MicroRNA Pathway in Mammalian Oocytes. Nucleic Acids Res. 2020, 48, 8050–8062. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Amikura, R.; Hanyu, K.; Kobayashi, S. Me31B Silences Translation of Oocyte-Localizing RNAs through the Formation of Cytoplasmic RNP Complex during Drosophila Oogenesis. Development 2001, 128, 3233–3242. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Lasko, P. Translational Control in Oocyte Development. Cold Spring Harb. Perspect. Biol. 2011, 3, a002758. [Google Scholar] [CrossRef] [PubMed]
- Christou-Kent, M.; Dhellemmes, M.; Lambert, E.; Ray, P.F.; Arnoult, C. Diversity of RNA-Binding Proteins Modulating Post-Transcriptional Regulation of Protein Expression in the Maturing Mammalian Oocyte. Cells 2020, 9, 662. [Google Scholar] [CrossRef]
- Zheng, P.; Patel, B.; McMenamin, M.; Reddy, S.E.; Paprocki, A.M.; Schramm, R.D.; Latham, K.E. The Primate Embryo Gene Expression Resource: A Novel Resource to Facilitate Rapid Analysis of Gene Expression Patterns in Non-Human Primate Oocytes and Preimplantation Stage Embryos. Biol. Reprod. 2004, 70, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Mtango, N.R.; Potireddy, S.; Latham, K.E. Expression of MicroRNA Processing Machinery Genes in Rhesus Monkey Oocytes and Embryos of Different Developmental Potentials. Mol. Reprod. Dev. 2009, 76, 255–269. [Google Scholar] [CrossRef]
- Ma, J.; Flemr, M.; Stein, P.; Berninger, P.; Malik, R.; Zavolan, M.; Svoboda, P.; Schultz, R.M. MicroRNA Activity Is Suppressed in Mouse Oocytes. Curr. Biol. 2010, 20, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Baehner, L.; Moltzahn, F.; Melton, C.; Shenoy, A.; Chen, J.; Blelloch, R. MicroRNA Function Is Globally Suppressed in Mouse Oocytes and Early Embryos. Curr. Biol. 2010, 20, 271–277. [Google Scholar] [CrossRef]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer Is Essential for Mouse Development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Kataruka, S.; Kinterova, V.; Horvat, F.; Kulmann, M.I.R.; Kanka, J.; Svoboda, P. Physiologically Relevant MiRNAs in Mammalian Oocytes Are Rare and Highly Abundant. EMBO Rep. 2022, 23, e53514. [Google Scholar] [CrossRef]
- Orisaka, M.; Miyazaki, Y.; Shirafuji, A.; Tamamura, C.; Tsuyoshi, H.; Tsang, B.K.; Yoshida, Y. The Role of Pituitary Gonadotropins and Intraovarian Regulators in Follicle Development: A Mini-Review. Reprod. Med. Biol. 2021, 20, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, F.; Gerris, J.; Vansteelandt, S.; De Baerdemaeker, A.; Tilleman, K.; De Sutter, P. Delaying the Oocyte Maturation Trigger by One Day Leads to a Higher Metaphase II Oocyte Yield in IVF/ICSI: A Randomised Controlled Trial. Reprod. Biol. Endocrinol. 2014, 12, 31. [Google Scholar] [CrossRef]
- Alviggi, C.; Conforti, A.; Esteves, S.C.; Andersen, C.Y.; Bosch, E.; Bühler, K.; Ferraretti, A.P.; De Placido, G.; Mollo, A.; Fischer, R.; et al. Recombinant Luteinizing Hormone Supplementation in Assisted Reproductive Technology: A Systematic Review. Fertil. Steril. 2018, 109, 644–664. [Google Scholar] [CrossRef] [PubMed]
- Conforti, A.; Esteves, S.C.; Di Rella, F.; Strina, I.; De Rosa, P.; Fiorenza, A.; Zullo, F.; De Placido, G.; Alviggi, C. The Role of Recombinant LH in Women with Hypo-Response to Controlled Ovarian Stimulation: A Systematic Review and Meta-Analysis. Reprod. Biol. Endocrinol. 2019, 17, 18. [Google Scholar] [CrossRef]
- Levi-Setti, P.E.; Zerbetto, I.; Baggiani, A.; Zannoni, E.; Sacchi, L.; Smeraldi, A.; Morenghi, E.; De Cesare, R.; Drovanti, A.; Santi, D. An Observational Retrospective Cohort Trial on 4,828 IVF Cycles Evaluating Different Low Prognosis Patients Following the POSEIDON Criteria. Front. Endocrinol. 2019, 10, 282. [Google Scholar] [CrossRef]
- Arvis, P.; Massin, N.; Lehert, P. Effect of Recombinant LH Supplementation on Cumulative Live Birth Rate Compared with FSH Alone in Poor Ovarian Responders: A Large, Real-World Study. Reprod. Biomed. Online 2021, 42, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Gatta, V.; Tatone, C.; Ciriminna, R.; Vento, M.; Franchi, S.; d’Aurora, M.; Sperduti, S.; Cela, V.; Borzì, P.; Palermo, R.; et al. Gene Expression Profiles of Cumulus Cells Obtained from Women Treated with Recombinant Human Luteinizing Hormone + Recombinant Human Follicle-Stimulating Hormone or Highly Purified Human Menopausal Gonadotropin versus Recombinant Human Follicle-Stimulating Hormone Alone. Fertil. Steril. 2013, 99, 2000–2008.e1. [Google Scholar] [CrossRef] [PubMed]
- Tatone, C.; Benedetti, E.; Vitti, M.; Di Emidio, G.; Ciriminna, R.; Vento, M.E.; Cela, V.; Borzì, P.; Carta, G.; Lispi, M.; et al. Modulating Intrafollicular Hormonal Milieu in Controlled Ovarian Stimulation: Insights from PPAR Expression in Human Granulosa Cells. J. Cell Physiol. 2016, 231, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Artini, P.G.; Tatone, C.; Sperduti, S.; D’Aurora, M.; Franchi, S.; Di Emidio, G.; Ciriminna, R.; Vento, M.; Di Pietro, C.; Stuppia, L.; et al. Cumulus Cells Surrounding Oocytes with High Developmental Competence Exhibit Down-Regulation of Phosphoinositol 1,3 Kinase/Protein Kinase B (PI3K/AKT) Signalling Genes Involved in Proliferation and Survival. Hum. Reprod. 2017, 32, 2474–2484. [Google Scholar] [CrossRef]
- Vigo, F.M.; Fraietta, R.; Rodrigues, F.; Carvalho, C.V.; Bonetti, T.C.S.; Motta, E.L.A. Cumulus Cell MicroRNA Expression When LH Is Added to the Ovarian Stimulation Protocol: A Pilot Study. Reprod. BioMedicine Online 2021, 43, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Dell’Aversana, C.; Cuomo, F.; Longobardi, S.; D’Hooghe, T.; Caprio, F.; Franci, G.; Santonastaso, M.; Colacurci, N.; Barone, S.; Pisaturo, V.; et al. Age-Related MiRNome Landscape of Cumulus Oophorus Cells during Controlled Ovarian Stimulation Protocols in IVF Cycles. Hum. Reprod. 2021, 36, 1310–1325. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.H.; Nunes, S.G.; Franchi, F.F.; Giroto, A.B.; Fontes, P.K.; Pinheiro, V.G.; Castilho, A.C.S. Expression of Bta-MiR-222 and LHCGR in Bovine Cultured Granulosa Cells: Impact of Follicle Deviation and Regulation by FSH/Insulin in Vitro. Theriogenology 2022, 182, 71–77. [Google Scholar] [CrossRef]
- Konstantinidou, F.; Budani, M.C.; Sarra, A.; Stuppia, L.; Tiboni, G.M.; Gatta, V. Impact of Cigarette Smoking on the Expression of Oxidative Stress-Related Genes in Cumulus Cells Retrieved from Healthy Women Undergoing IVF. Int. J. Mol. Sci. 2021, 22, 13147. [Google Scholar] [CrossRef] [PubMed]
- Mochtar, M.H.; Danhof, N.A.; Ayeleke, R.O.; Veen, F.V.d.; van Wely, M. Recombinant Luteinizing Hormone (RLH) and Recombinant Follicle Stimulating Hormone (RFSH) for Ovarian Stimulation in IVF/ICSI Cycles. Cochrane Database Syst. Rev. 2017, 5, 1465–1858. [Google Scholar] [CrossRef]
- Kaur, S.; Kurokawa, M. Regulation of Oocyte Apoptosis: A View from Gene Knockout Mice. Int. J. Mol. Sci. 2023, 24, 1345. [Google Scholar] [CrossRef] [PubMed]
- Zanjirband, M.; Hodayi, R.; Safaeinejad, Z.; Nasr-Esfahani, M.H.; Ghaedi-Heydari, R. Evaluation of the P53 Pathway in Polycystic Ovarian Syndrome Pathogenesis and Apoptosis Enhancement in Human Granulosa Cells through Transcriptome Data Analysis. Sci. Rep. 2023, 13, 11648. [Google Scholar] [CrossRef]
- Luan, Y.; Xu, P.; Yu, S.-Y.; Kim, S.-Y. The Role of Mutant P63 in Female Fertility. Int. J. Mol. Sci. 2021, 22, 8968. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, H.; Hirota, Y.; Saito-Fujita, T.; Tanaka, T.; Shimizu-Hirota, R.; Harada, M.; Akaeda, S.; Hiraoka, T.; Matsuo, M.; Matsumoto, L.; et al. Mdm2-P53-SF1 Pathway in Ovarian Granulosa Cells Directs Ovulation and Fertilization by Conditioning Oocyte Quality. FASEB J. 2019, 33, 2610–2620. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, Y.; Tsuno, S.; Ping, B.; Ashizaki, T.; Nakashima, M.; Miura, K.; Miura, Y.; Yamashita, T.; Hasegawa, J.; Miura, N. Hsa-MiR-520d-5p Promotes Survival in Human Dermal Fibroblasts Exposed to a Lethal Dose of UV Irradiation. npj Aging Mech. Dis. 2016, 2, 16029. [Google Scholar] [CrossRef]
- Chen, W.; Li, X. MiR-222-3p Promotes Cell Proliferation and Inhibits Apoptosis by Targeting PUMA (BBC3) in Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820922558. [Google Scholar] [CrossRef]
- Nie, W.; Huang, X.; Zhao, L.; Wang, T.; Zhang, D.; Xu, T.; Du, L.; Li, Y.; Zhang, W.; Xiao, F.; et al. Exosomal MiR-17–92 Derived from Human Mesenchymal Stem Cells Promotes Wound Healing by Enhancing Angiogenesis and Inhibiting Endothelial Cell Ferroptosis. Tissue Cell 2023, 83, 102124. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, T.; Huang, Q.-K.; Pu, M.; Sun, W.; Zhang, Z.-C.; Ling, R.; Tao, K.-S. MicroRNA-548a-5p Promotes Proliferation and Inhibits Apoptosis in Hepatocellular Carcinoma Cells by Targeting Tg737. World J. Gastroenterol. 2016, 22, 5364–5373. [Google Scholar] [CrossRef]
- Jiang, W.; Li, T.; Wang, J.; Jiao, R.; Shi, X.; Huang, X.; Ji, G. MiR-140-3p Suppresses Cell Growth and Induces Apoptosis In Colorectal Cancer By Targeting PD-L1. OncoTargets Ther. 2019, 12, 10275–10285. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, X.; Mu, H.; Mei, Q.; Liu, Y.; Min, Z.; Zhang, L.; Su, P.; Xiang, W. Mir-484 Contributes to Diminished Ovarian Reserve by Regulating Granulosa Cell Function via YAP1-Mediated Mitochondrial Function and Apoptosis. Int. J. Biol. Sci. 2022, 18, 1008–1021. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, J.; Li, H.; Mu, H.; Zeng, L.; Cai, S.; Su, P.; Li, H.; Zhang, L.; Xiang, W. MiR-484 Mediates Oxidative Stress-Induced Ovarian Dysfunction and Promotes Granulosa Cell Apoptosis via SESN2 Downregulation. Redox Biol. 2023, 62, 102684. [Google Scholar] [CrossRef] [PubMed]
- Saadia, Z. Follicle Stimulating Hormone (LH: FSH) Ratio in Polycystic Ovary Syndrome (PCOS) - Obese vs. Non-Obese Women. Med. Arch. 2020, 74, 289–293. [Google Scholar] [CrossRef]
- Hossain, M.M.; Cao, M.; Wang, Q.; Kim, J.Y.; Schellander, K.; Tesfaye, D.; Tsang, B.K. Altered Expression of MiRNAs in a Dihydrotestosterone-Induced Rat PCOS Model. J. Ovarian Res. 2013, 6, 36. [Google Scholar] [CrossRef]
- Huang, X.; She, L.; Luo, X.; Huang, S.; Wu, J. MiR-222 Promotes the Progression of Polycystic Ovary Syndrome by Targeting P27 Kip1. Pathol. Res. Pract. 2019, 215, 918–923. [Google Scholar] [CrossRef]
- Donker, R.B.; Mouillet, J.F.; Chu, T.; Hubel, C.A.; Stolz, D.B.; Morelli, A.E.; Sadovsky, Y. The Expression Profile of C19MC MicroRNAs in Primary Human Trophoblast Cells and Exosomes. Mol. Hum. Reprod. 2012, 18, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.; Vento, M.E.; Ragusa, M.; Barbagallo, D.; La Ferlita, A.; Di Emidio, G.; Borzí, P.; Artini, P.G.; Scollo, P.; Tatone, C.; et al. MicroRNAs Are Stored in Human MII Oocyte and Their Expression Profile Changes in Reproductive Aging. Biol. Reprod. 2016, 95, 131. [Google Scholar] [CrossRef]
- Battaglia, R.; Vento, M.E.; Borzì, P.; Ragusa, M.; Barbagallo, D.; Arena, D.; Purrello, M.; Di Pietro, C. Non-Coding RNAs in the Ovarian Follicle. Front. Genet. 2017, 8, 57. [Google Scholar] [CrossRef]
- Zhang, D.; Lv, J.; Tang, R.; Feng, Y.; Zhao, Y.; Fei, X.; Chian, R.; Xie, Q. Association of Exosomal MicroRNAs in Human Ovarian Follicular Fluid with Oocyte Quality. Biochem. Biophys. Res. Commun. 2021, 534, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Son, G.-H.; Kim, Y.; Lee, J.J.; Lee, K.-Y.; Ham, H.; Song, J.-E.; Park, S.T.; Kim, Y.-H. MicroRNA-548 Regulates High Mobility Group Box 1 Expression in Patients with Preterm Birth and Chorioamnionitis. Sci. Rep. 2019, 9, 19746. [Google Scholar] [CrossRef]
- Pais, H.; Nicolas, F.E.; Soond, S.M.; Swingler, T.E.; Clark, I.M.; Chantry, A.; Moulton, V.; Dalmay, T. Analyzing MRNA Expression Identifies Smad3 as a MicroRNA-140 Target Regulated Only at Protein Level. RNA 2010, 16, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Paria, B.C.; Jones, K.L.; Flanders, K.C.; Dey, S.K. Localization and Binding of Transforming Growth Factor-Beta Isoforms in Mouse Preimplantation Embryos and in Delayed and Activated Blastocysts. Dev. Biol. 1992, 151, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Mummery, C.L. Transforming Growth Factor Beta and Mouse Development. Microsc. Res. Tech. 2001, 52, 374–386. [Google Scholar] [CrossRef]
- Liu, W.-M.; Pang, R.T.K.; Cheong, A.W.Y.; Ng, E.H.Y.; Lao, K.; Lee, K.-F.; Yeung, W.S.B. Involvement of MicroRNA Lethal-7a in the Regulation of Embryo Implantation in Mice. PLoS ONE 2012, 7, e37039. [Google Scholar] [CrossRef]
- ElKhouly, A.M.; Youness, R.A.; Gad, M.Z. MicroRNA-486-5p and microRNA-486-3p: Multifaceted pleiotropic mediators in oncological and non-oncological conditions. Non-Coding RNA Res. 2020, 5, 11–21. [Google Scholar] [CrossRef]
- Ye, H.; Yu, X.; Xia, J.; Tang, X.; Tang, L.; Chen, F. MiR-486-3p targeting ECM1 represses cell proliferation and metastasis in cervical cancer. Biomed. Pharmacother. 2016, 80, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhang, X.; Yang, Q.; Wang, J.; He, Y.; Sun, Z.; Zhang, H.; Wang, J. Aberrant Placental Villus Expression of miR-486-3p and miR-3074-5p in Recurrent Miscarriage Patients and Uterine Expression of These MicroRNAs during Early Pregnancy in Mice. Gynecol. Obstet. Investig. 2015, 81, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Hill, B.G.; Benavides, G.A.; Lancaster, J.R.; Ballinger, S.; Dell’Italia, L.; Zhang, J.; Darley-Usmar, V.M. Integration of Cellular Bioenergetics with Mitochondrial Quality Control and Autophagy. Biol. Chem. 2012, 393, 1485–1512. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, A.; Kim, B.; Yeh, J. Luteinizing Hormone Action in Human Oocyte Maturation and Quality: Signaling Pathways, Regulation, and Clinical Impact. Reprod. Sci. 2020, 27, 1223–1252. [Google Scholar] [CrossRef] [PubMed]
- Marchiani, S.; Tamburrino, L.; Benini, F.; Pallecchi, M.; Bignozzi, C.; Conforti, A.; Alviggi, C.; Vignozzi, L.; Danza, G.; Pellegrini, S.; et al. LH Supplementation of Ovarian Stimulation Protocols Influences Follicular Fluid Steroid Composition Contributing to the Improvement of Ovarian Response in Poor Responder Women. Sci. Rep. 2020, 10, 12907. [Google Scholar] [CrossRef] [PubMed]


| r-hFSH | r-hFSH + r-hLH | p Value | |
|---|---|---|---|
| No. of patients | 7 | 6 | NS |
| Age (y) | 33.7 ± 3.1 | 35.3 ± 2.1 | NS |
| FSH (IU/mL) | 7.7 ± 2.1 | 8.1 ± 1.8 | NS |
| LH (IU/mL) | 5.7 ± 2.3 | 5.1 ± 2.1 | NS |
| AMH (ng/mL) | 1.9 ± 0.9 | 1.7 ± 0.7 | NS |
| AFC | 13.7 ± 2.5 | 12.9 ± 2.3 | NS |
| No. supernumerary oocytes MII | 24 | 20 | NS |
| No. of microRNAs | r-hFSH | r-hFSH + r-hLH |
|---|---|---|
| 1 | hsa-miR-548c | hsa-miR-548c |
| 2 | hsa-miR-222 | hsa-miR-222 |
| 3 | hsa-miR-484 | hsa-miR-484 |
| 4 | hsa-miR-539 | hsa-miR-486-3p |
| 5 | hsa-miR-636 | hsa-miR-17 |
| 6 | hsa-miR-17 | mmu-miR-140 |
| 7 | hsa-miR-130a | hsa-miR-520d-5p |
| 8 | mmu-miR-140 | hsa-miR-518d |
| 9 | hsa-miR-520b | hsa-miR-548a |
| 10 | hsa-let-7b | hsa-miR-627 |
| 11 | hsa-miR-106a | - |
| 12 | hsa-miR-520d-5p | - |
| 13 | hsa-miR-518d | - |
| 14 | hsa-miR-548a | - |
| 15 | hsa-miR-628-5p | - |
| 16 | hsa-miR-525-3p | - |
| 17 | hsa-miR-627 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konstantinidou, F.; Placidi, M.; Di Emidio, G.; Stuppia, L.; Tatone, C.; Gatta, V.; Artini, P.G. Maternal MicroRNA Profile Changes When LH Is Added to the Ovarian Stimulation Protocol: A Pilot Study. Epigenomes 2023, 7, 25. https://doi.org/10.3390/epigenomes7040025
Konstantinidou F, Placidi M, Di Emidio G, Stuppia L, Tatone C, Gatta V, Artini PG. Maternal MicroRNA Profile Changes When LH Is Added to the Ovarian Stimulation Protocol: A Pilot Study. Epigenomes. 2023; 7(4):25. https://doi.org/10.3390/epigenomes7040025
Chicago/Turabian StyleKonstantinidou, Fani, Martina Placidi, Giovanna Di Emidio, Liborio Stuppia, Carla Tatone, Valentina Gatta, and Paolo Giovanni Artini. 2023. "Maternal MicroRNA Profile Changes When LH Is Added to the Ovarian Stimulation Protocol: A Pilot Study" Epigenomes 7, no. 4: 25. https://doi.org/10.3390/epigenomes7040025
APA StyleKonstantinidou, F., Placidi, M., Di Emidio, G., Stuppia, L., Tatone, C., Gatta, V., & Artini, P. G. (2023). Maternal MicroRNA Profile Changes When LH Is Added to the Ovarian Stimulation Protocol: A Pilot Study. Epigenomes, 7(4), 25. https://doi.org/10.3390/epigenomes7040025









