- Review
Genetic–Epigenetic Interplay in Epilepsy: Pathways, Biomarkers, and Epigenome-Targeted Therapies
- Andra-Giorgiana Zaruha,
- Patricia Codreanu and
- Viorica-Elena Rădoi
- + 11 authors
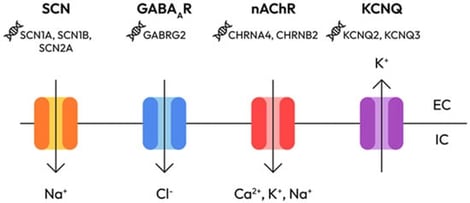
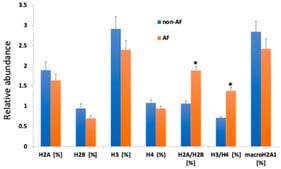
Epilepsy is a heterogeneous neurological disorder with a strong genetic basis, yet recent evidence underscores the critical role of epigenetic mechanisms in its pathogenesis. This review synthesizes current knowledge on how chromatin remodeling, histone modifications, DNA methylation, and transcriptional regulation intersect with classical channelopathies and signaling pathways. We emphasize how epigenetic dysregulation contributes to neuronal excitability and network plasticity, particularly through interactions with mTOR, PI3K-AKT, and GABAergic signaling cascades. The convergence of genetic mutations and epigenetic modifications creates a dynamic landscape in which environmental factors can modify gene expression and contribute to the development of epilepsy. Emerging therapeutic strategies—including epigenetic drugs (HDAC inhibitors, DNMT inhibitors), CRISPR/dCas9-based epigenome editing, and multi-omics approaches—offer promising avenues for precision medicine. This review provides a comprehensive synthesis of genetic and epigenetic mechanisms in epilepsy, examining how these layers interact to produce disease phenotypes and discussing the therapeutic implications of this multilayered regulation.
10 February 2026





![HSP70-1 is important for MSUD. In a normal cross, Neurospora produces American football-shaped spores. In a round spore (r+)-unpaired cross (i.e., r+ × r∆), r+ is silenced, and predominantly round spores are produced (i.e., 0.38% football; cross 1). In an hsp70-1-null background, the silencing of an unpaired r+ gene becomes deficient, and significantly more normal spores are produced (i.e., 77.8% football, cross 2; p < 0.001). Suppression of silencing is nearly complete (i.e., 98.9% football, cross 3) when the cross is lacking SAD-5 (a protein required for siRNA production) [41]. An error bar indicates the standard deviation among 24 replicates. +, wild type at pertinent loci. Crosses: (1) F9-37 × P3-08. (2) F9-18 × P27-55. (3) F5-36 × P17-70.](https://mdpi-res.com/cdn-cgi/image/w=281,h=192/https://mdpi-res.com/epigenomes/epigenomes-10-00007/article_deploy/html/images/epigenomes-10-00007-g001-550.jpg)