3.1. Statistical Model Selection for Predicting IM Based on TNSC
In this study, we first match the DNA methylation data downloaded from NCBI (
https://www.ncbi.nlm.nih.gov/geo/, GSE103186, accessed on 23 January 2024) with the tissue samples listed in Table S3 in [
5] (
https://www.cell.com/cancer-cell/, accessed on 18 January 2024). Among the 134 tissue samples collected at the antrum site [
5], there are 10 samples lacking DNA methylation profiles. We use the remaining 124 samples for our analysis. We then compute the TNSC values for the 124 samples using their DNA methylation data, as described in
Section 2.1. The R codes for computing TNSC are accessible online (
https://zenodo.org/records/2632938, epiTOC2.R, accessed on 15 January 2024) as indicated by [
12]. In this section, we consider the multinomial mixed-link model as described in
Section 2.2, and use the computed TNSC as the only covariate to predict the risk level of IM in three categories (Normal, MIM, and IM). For each of
models, the optimal link functions for
, respectively, along with their corresponding AIC and BIC values, are listed in
Table 1 (see
Appendix A for the AIC and BIC values of all link combinations).
According to
Table 1, the best multinomial mixed-link model with the lowest AIC overall in this case, called Model 1, is a cumulative po model with loglog and logit links for
(Normal) and
(MIM), respectively. Note that by default
(IM) is treated as the baseline category. The fitted Model 1 is provided in (
8), where
is the computed TNSC value for the
ith tissue sample.
In (
8), the estimated coefficient of
is
, which is fairly small. To test whether the effect of TNSC is significant in predicting IM, we obtain its
confidence interval
, which does not contain zero. Actually, the corresponding
p-value of its significance test is less than
. As a conclusion, the effect of TNSC is statistically significant in predicting the risk level of IM.
To further check whether Model 1 outperforms the traditional statistical models, as described in
Section 2.3, we run a ten-fold cross-validation and compare its cross-entropy loss against other models. For illustration purposes, we choose the baseline-category logit model with npo (also known as the multiclass logistic regression model) and the cumulative logit model with npo (one of the most popular models for ordinal responses) as the alternative models. As for other models, including multinomial logit models and probit models, the conclusions are similar (see
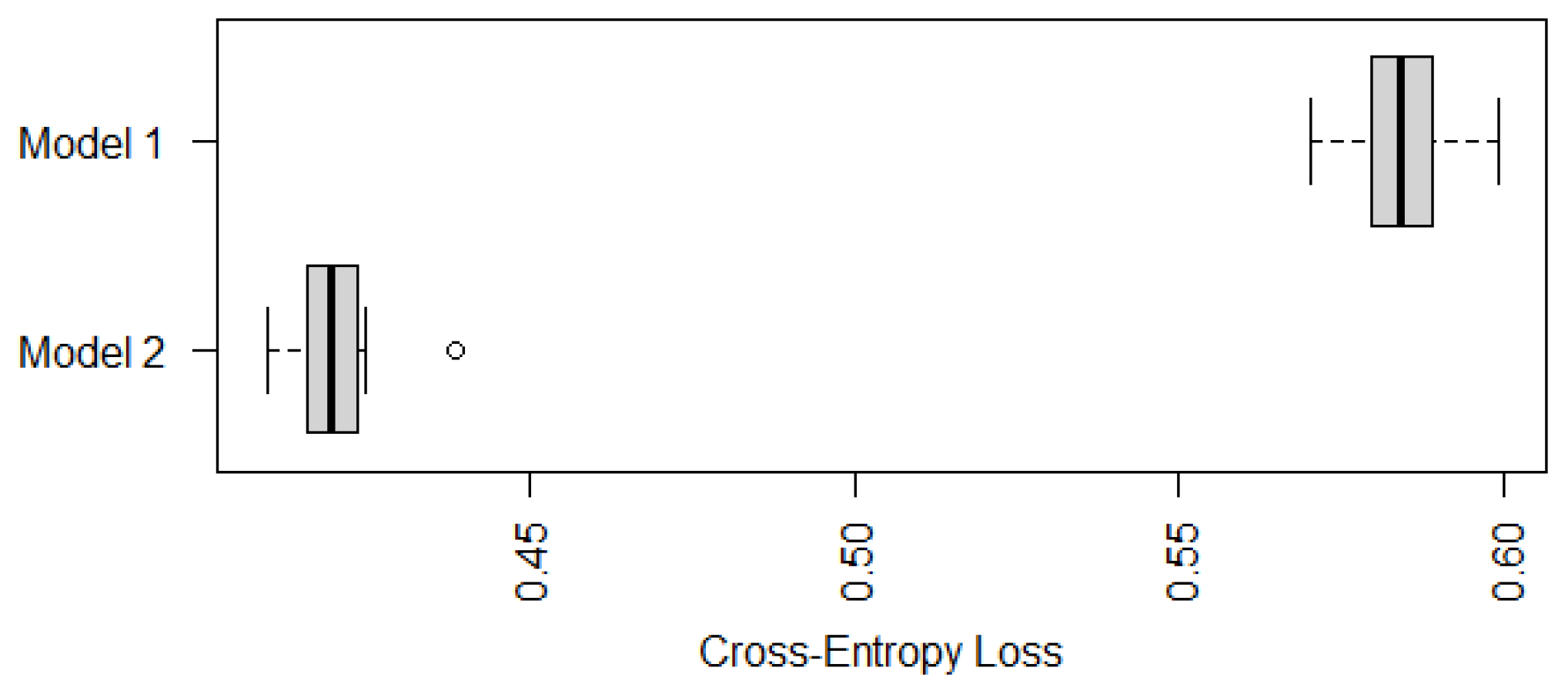
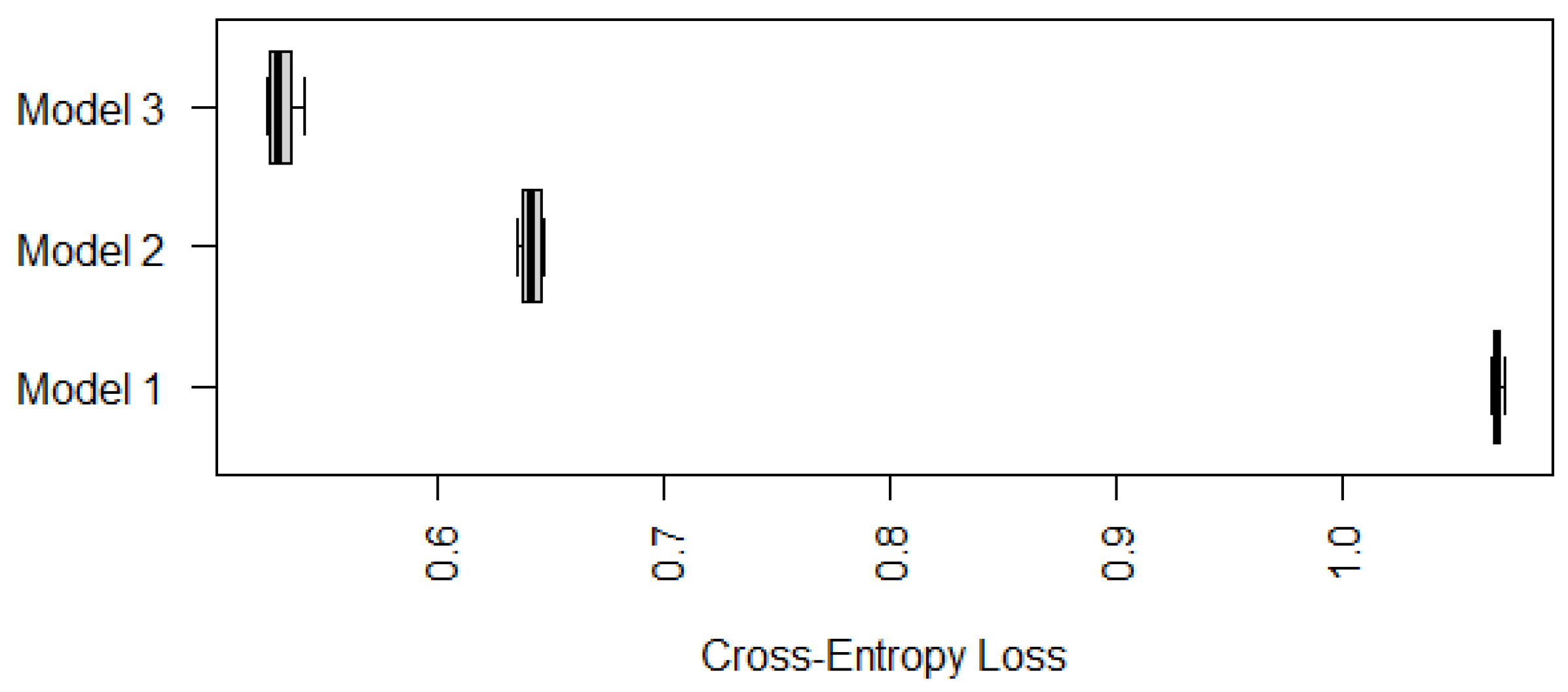
Appendix A). To avoid misleading conclusions relying on a particular partition, we randomly generate ten partitions and compute their corresponding CE values. The boxplots of the resulting ten CE values are provided in
Figure 1, which shows that the CE values of Model 1 seem to be much lower than those values of the other two models. Although we only run ten random partitions due to computational intensity, our one-sided paired
t-tests based on the ten CE values show that the improvements of Model 1 are significant. The
p-values of the
t-tests for comparing Model 1 against the baseline npo model and the cumulative npo model displayed in
Figure 1 are
and
, respectively. That is, the recommended cumulative po model with loglog and logit links significantly outperforms the two multinomial logistic models that are commonly used in practice.
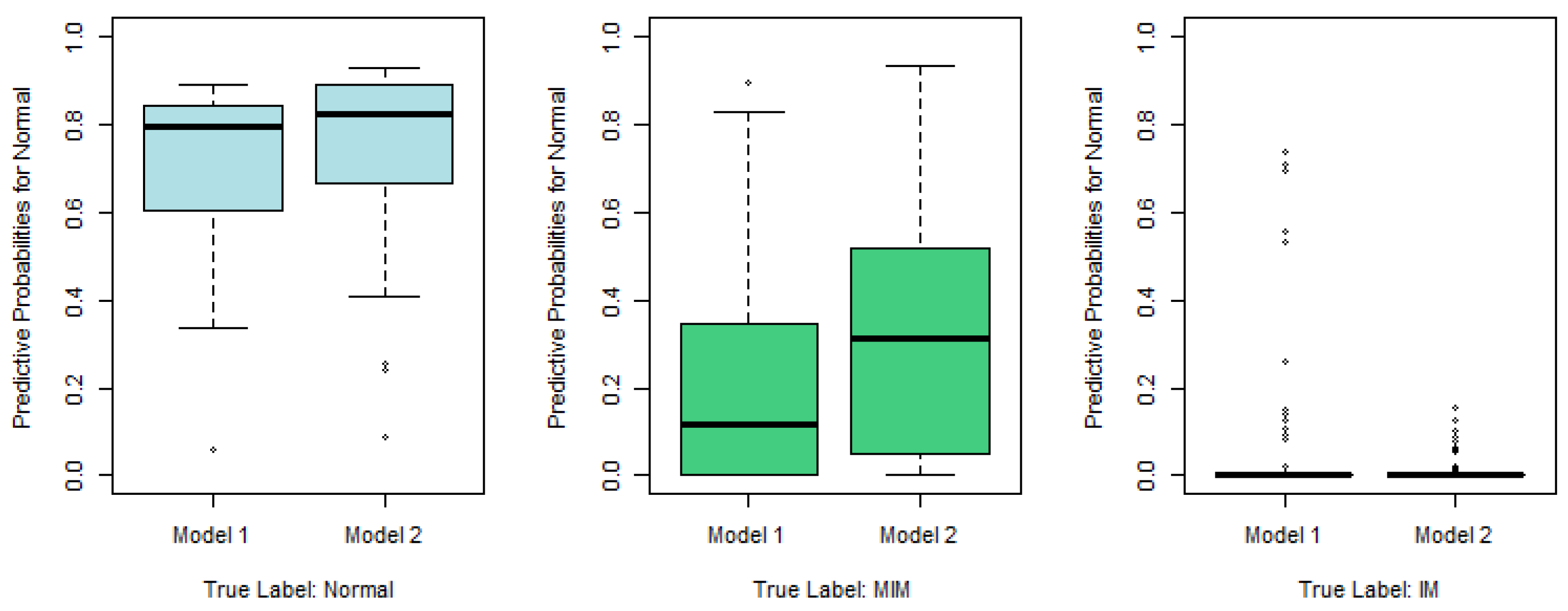
To show how well Model 1 works, we plot in
Figure 2 the predictive probabilities
against the true response labels,
, respectively.
According to
Figure 2, the recommended Model 1 works reasonably well. For examples, in the left panel, we plot
, which is the predictive probability that the
ith tissue sample belongs to Normal, against its true response label. If the true label is Normal, the left boxplot in the left panel of
Figure 2, which is apparently higher than the other two boxplots in the same panel, indicates that the corresponding tissue sample tends to be predicted as Normal as well. Similarly, in the right panel,
, the predictive probability that the sample belongs to IM, is plotted, and the significantly higher boxplot to the right indicates that the sample with true label IM tends to be predicted as IM as well. Nevertheless, the middle panel, which plots the predictive probabilities for MIM, indicates that the MIM class is not so different from Normal or IM, and thus is more difficult to predict correctly.
3.2. Statistical Model Selection for Predicting IM Based on TNSC and Gastric Atrophy
In this section, we show that when additional information, such as the status of gastric atrophy, is available, the prediction accuracy of the IM risk level can be significantly improved.
In this study, the status of gastric atrophy is a 5-class categorical variable (see Table S3 in [
5]), namely, Marked, Moderate, Mild, Negative, and Unknown. In our regression analysis involving the status of gastric atrophy, we replace it with four dummy variables:
,
,
, and
. Each dummy variable is binary, taking a value of either 1 or 0, with at most one variable allowed to be 1 for any given sample. For instance, a configuration of
indicates a mild gastric atrophy status for the
ith sample,
indicates a moderate gastric atrophy status, whereas
indicates a marked status, that is, the baseline status. Similarly to
Table 1, we list the optimal link functions for
, respectively, along with their AIC and BIC values, in
Table 2.
With the presence of gastric atrophy, the best multinomial mixed-link model, called Model 2, is an adjacent-categories logit model with po, which is different from the type of Model 1 with TNSC only (see
Section 3.1). Since its AIC value,
, is much less than
in
Table 1, Model 2 is expected to outperform Model 1 significantly in terms of prediction accuracy (see [
36] for more discussion on AIC differences). The fitted Model 2 is provided in (
9).
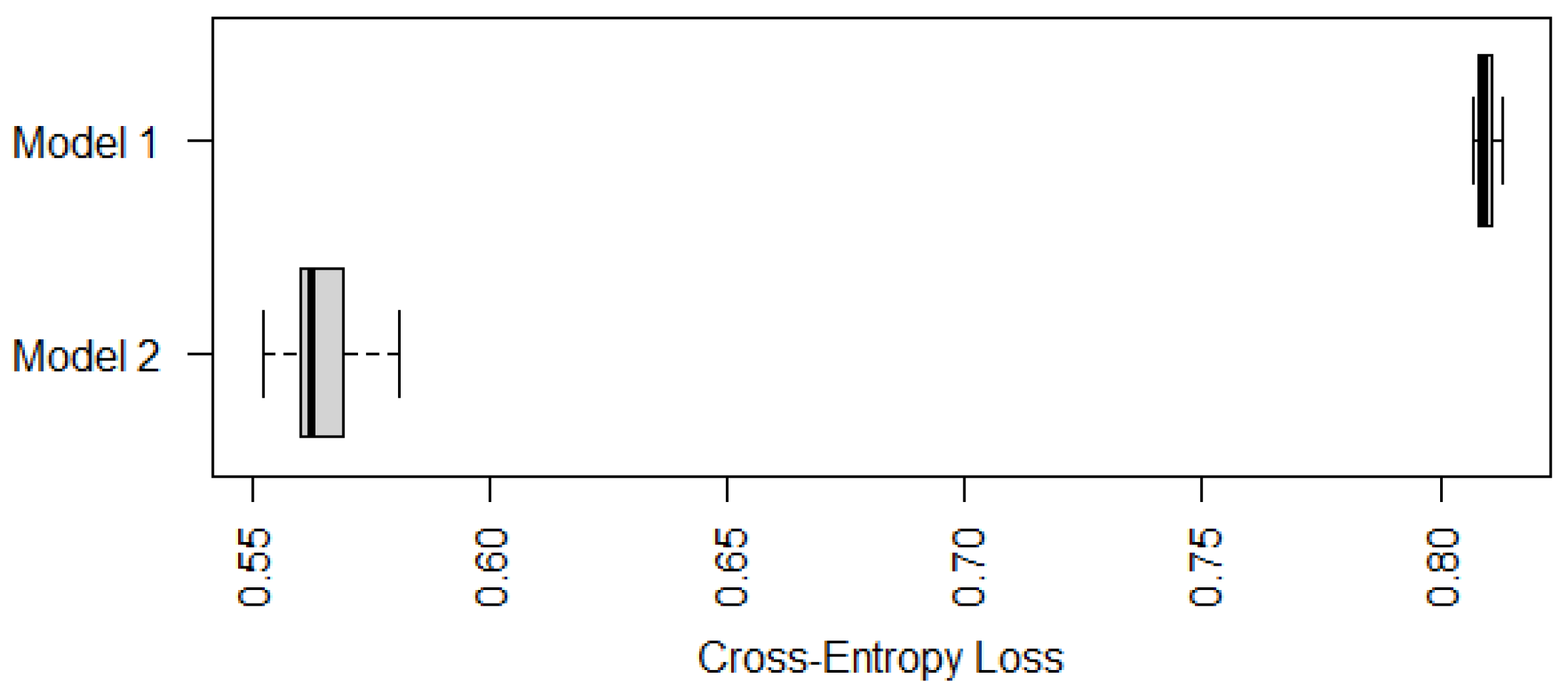
Similarly to
Figure 1, we compare in
Figure 3 the cross-entropy loss of two recommended models shown in (
8) (Model 1) and (
9) (Model 2). It is not surprising that Model 2 with both TNSC and gastric atrophy as predictors has a significantly smaller cross-entropy loss, which implies that the status of gastric atrophy is informative in predicting the risk level of IM.
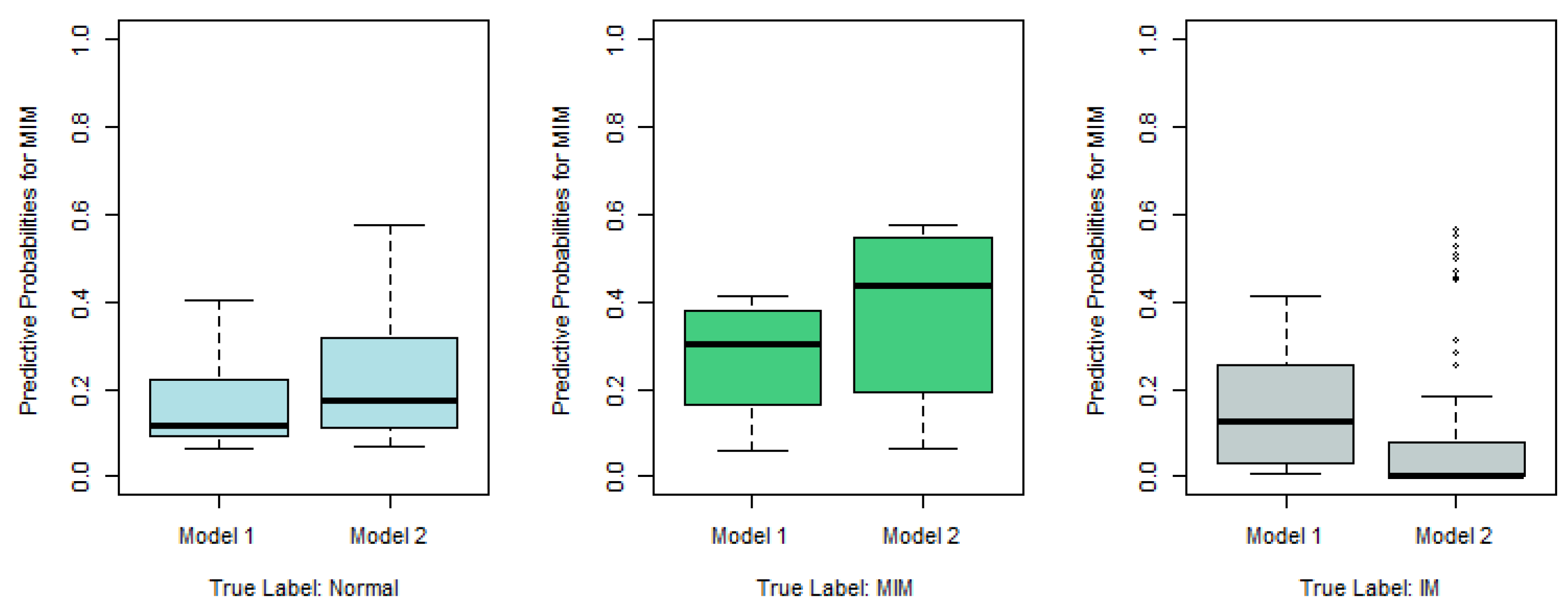
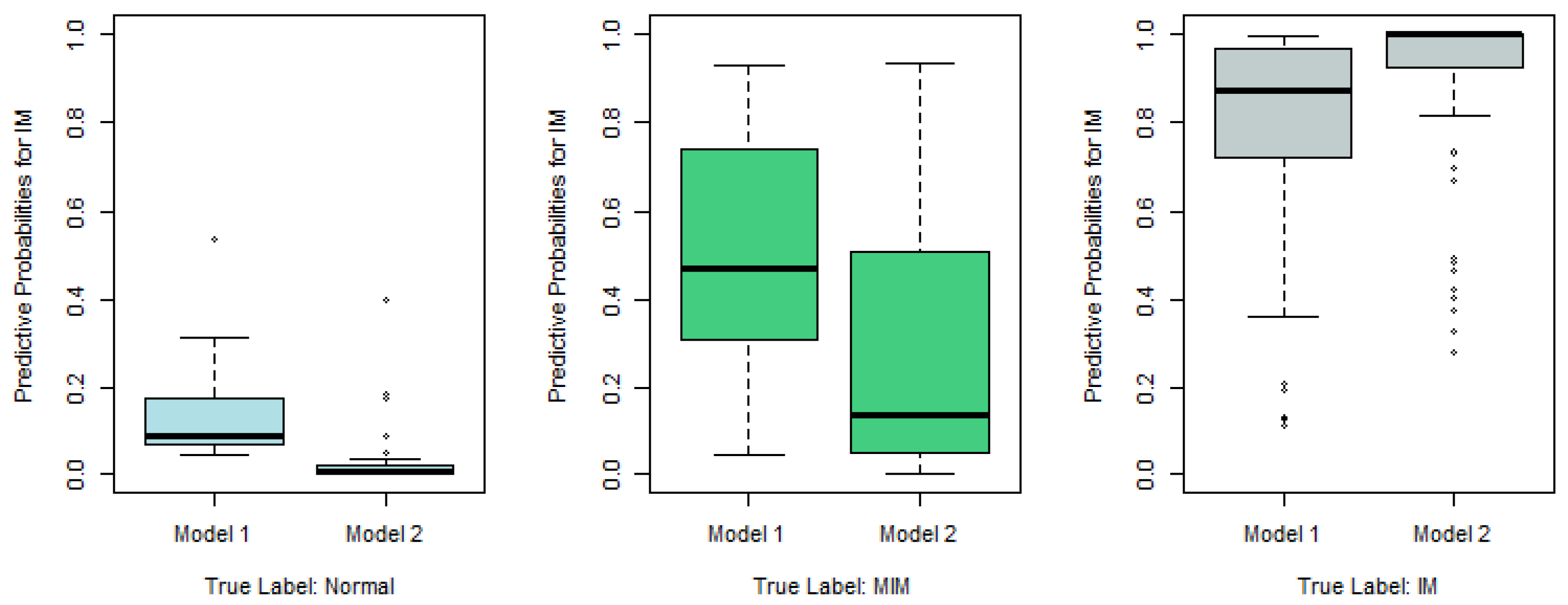
Similarly to
Figure 2, we plot the predictive probabilities based on Model 1 and Model 2 against the true IM labels in
Figure 4,
Figure 5 and
Figure 6. When the true IM label matches the predictive label, such as the left panel in
Figure 4, the middle panel in
Figure 5, and the right panel of
Figure 6, Model 2 tends to provide a higher predictive probability than Model 1, which shows that overall Model 2 outperforms Model 1.
3.3. Statistical Model Selection after Removing Unknown and Marked Categories
Among the 124 samples considered in this study, there are only 3 cases with “Marked” status of gastric atrophy, and there are 23 cases with “Unknown” status, which is not informative. In this section, we consider the best multinomial mixed-link model for the 98 cases after removing the samples that belong to Marked or Unknown categories.
In this section, the status of gastric atrophy is a three-class categorical variable restricted to the 98 samples. Similarly to Model 2 in
Section 3.2, we replace the status of gastric atrophy with two dummy variables (
,
). More specifically, (
,
) = (1,0) stands for mild status, (0,1) for moderate status, and (0,0) for negative status representing the baseline. Similarly to
Table 1 and
Table 2, we provide in
Table 3 the optimal choices of link functions for each type of multinomial model. According to
Table 3, the best multinomial mixed-link model for this scenario is an adjacent-categories po model with probit links for both
. We call it Model 3 and list its fitted model in (
10).
To compare the performance of Model 3 with Model 1 and Model 2, we use the cross-entropy loss based on ten-fold cross-validations similarly to
Section 3.1 and
Section 3.2. Since Model 3 cannot be applied to cases with marked or unknown status of gastric atrophy, we compare the performance of the three models on samples with mild, moderate, or negative status of gastric atrophy only. Their boxplots of cross-entropy loss based on ten random partitions for ten-fold cross-validations are displayed in
Figure 7.
According to
Figure 7, Model 3 has a significantly smaller (average) cross-entropy loss compared with Model 1 and Model 2, in terms of predicting IM for individuals whose gastric atrophy statuses are negative, mild or moderate. Nevertheless, Models 1 and 2 are still useful since they can be applied to cases with marked or unknown status of gastric atrophy as well.