Stress and DNA Methylation of Blood Leukocytes among Pregnant Latina Women
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Methylation Analysis
4.2. Stress Candidate Gene Selection
4.3. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- United States Census Bureau. Quick Facts. Available online: https://www.census.gov/quickfacts/fact/table/US/PST045221 (accessed on 8 July 2022).
- Osterman, M.; Hamilton, B.; Martin, J.A.; Driscoll, A.K.; Valenzuela, C.P. Births: Final Data for 2020. Natl. Vital. Stat. Rep. 2021, 70, 1–50. [Google Scholar] [PubMed]
- Dunkel Schetter, C.; Niles, A.N.; Guardino, C.M.; Khaled, M.; Kramer, M.S. Demographic, Medical, and Psychosocial Predictors of Pregnancy Anxiety. Paediatr. Perinat. Epidemiol. 2016, 30, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Alcalá, H.E.; Albert, S.L.; Trabanino, S.K.; Garcia, R.E.; Glik, D.C.; Prelip, M.L.; Ortega, A.N. Access to and Use of Health Care Services Among Latinos in East Los Angeles and Boyle Heights. Fam. Community Health 2016, 39, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, A.; McCormick, D. The Affordable Care Act: Implications for health-care equity. Lancet 2017, 389, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Barcelona de Mendoza, V.; Harville, E.; Theall, K.; Buekens, P.; Chasan-Taber, L. Acculturation and Adverse Birth Outcomes in a Predominantly Puerto Rican Population. Matern. Child Health J. 2016, 20, 1151–1160. [Google Scholar] [CrossRef]
- Kominiarek, M.A.; Cordero, C.; Stuebe, A.M.; Simon, M.; Evenson, K.R.; Perreira, K.M.; Gallo, L.C.; Castañeda, S.F.; Potter, J.E.; Wu, D.; et al. Pre-pregnancy Health Behaviors and Gestational Weight Gain Among Hispanic/Latino Women: Hispanic Community Health Study/Study of Latinos. Matern. Child Health J. 2021, 25, 2002–2013. [Google Scholar] [CrossRef]
- Howell, E.A.; Egorova, N.N.; Janevic, T.; Brodman, M.; Balbierz, A.; Zeitlin, J.; Hebert, P.L. Race and Ethnicity, Medical Insurance, and Within-Hospital Severe Maternal Morbidity Disparities. Obstet. Gynecol. 2020, 135, 285–293. [Google Scholar] [CrossRef]
- Casagrande, S.S.; Linder, B.; Cowie, C.C. Prevalence of gestational diabetes and subsequent Type 2 diabetes among U.S. women. Diabetes Res. Clin. Pract. 2018, 141, 200–208. [Google Scholar] [CrossRef]
- Campos, C.L.; Rodriguez, C.J. High blood pressure in Hispanics in the United States: A review. Curr. Opin. Cardiol. 2019, 34, 350–358. [Google Scholar] [CrossRef]
- Chalfun, G.; Reis, M.M.; de Oliveira, M.B.G.; de Araújo Brasil, A.; Dos Santos Salú, M.; da Cunha, A.; Prata-Barbosa, A.; de Magalhães-Barbosa, M.C. Perinatal stress and methylation of the NR3C1 gene in newborns: Systematic review. Epigenetics 2022, 17, 1003–1019. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, S.; Wang, G.; Hong, X.; Mallow, E.B.; Walker, S.O.; Pearson, C.; Heffner, L.; Zuckerman, B.; Wang, X. The combined association of psychosocial stress and chronic hypertension with preeclampsia. Am. J. Obstet. Gynecol. 2013, 209, 438.e1–438.e12. [Google Scholar] [CrossRef] [PubMed]
- Lara-Cinisomo, S.; Grewen, K.M.; Girdler, S.S.; Wood, J.; Meltzer-Brody, S. Perinatal Depression, Adverse Life Events, and Hypothalamic-Adrenal-Pituitary Axis Response to Cold Pressor Stress in Latinas: An Exploratory Study. Womens Health Issues 2017, 27, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Hamilton, B.E.; Osterman, M.J.K. Births in the United States, 2018. NCHS Data Brief 2019, 346, 1–8. [Google Scholar]
- Smith, A.K.; Conneely, K.N.; Kilaru, V.; Mercer, K.B.; Weiss, T.E.; Bradley, B.; Tang, Y.; Gillespie, C.F.; Cubells, J.F.; Ressler, K.J. Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. Am. J. Med. Genetics. Part B Neuropsychiatr. Genet. 2011, 156, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Hompes, T.; Izzi, B.; Gellens, E.; Morreels, M.; Fieuws, S.; Pexsters, A.; Schops, G.; Dom, M.; Van Bree, R.; Freson, K.; et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J. Psychiatr. Res. 2013, 47, 880–891. [Google Scholar] [CrossRef]
- Surkan, P.J.; Hong, X.; Zhang, B.; Nawa, N.; Ji, H.; Tang, W.Y.; Ji, Y.; Kimmel, M.C.; Wang, G.; Pearson, C.; et al. Can social support during pregnancy affect maternal DNA methylation? Findings from a cohort of African-Americans. Pediatr. Res. 2020, 88, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Torres-Salazar, Q.; Martínez-López, Y.; Reyes-Romero, M.; Pérez-Morales, R.; Sifuentes-Álvarez, A.; Salvador-Moysén, J. Differential Methylation in Promoter Regions of the Genes NR3C1 and HSP90AA1, Involved in the Regulation, and Bioavailability of Cortisol in Leukocytes of Women With Preeclampsia. Front. Med. 2020, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Kertes, D.A.; Kamin, H.S.; Hughes, D.A.; Rodney, N.C.; Bhatt, S.; Mulligan, C.J. Prenatal Maternal Stress Predicts Methylation of Genes Regulating the Hypothalamic-Pituitary-Adrenocortical System in Mothers and Newborns in the Democratic Republic of Congo. Child Dev. 2016, 87, 61–72. [Google Scholar] [CrossRef]
- Santos, H.P., Jr.; Adynski, H.; Harris, R.; Bhattacharya, A.; Incollingo Rodriguez, A.C.; Cali, R.; Yabar, A.T.; Nephew, B.C.; Murgatroyd, C. Biopsychosocial correlates of psychological distress in Latina mothers. J. Affect. Disord. 2021, 1, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Santos, H.P., Jr.; Nephew, B.C.; Bhattacharya, A.; Tan, X.; Smith, L.; Alyamani, R.A.S.; Martin, E.M.; Perreira, K.; Fry, R.C.; Murgatroyd, C. Discrimination exposure and DNA methylation of stress-related genes in Latina mothers. Psychoneuroendocrinology 2018, 98, 131–138. [Google Scholar] [CrossRef]
- Rowley, D.L.; Hogue, C.J.; Blackmore, C.A.; Ferre, C.D.; Hatfield-Timajchy, K.; Branch, P.; Atrash, H.K. Preterm delivery among African-American women: A research strategy. Am. J. Prev. Med. 1993, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Herman, J.P.; McKlveen, J.M.; Solomon, M.B.; Carvalho-Netto, E.; Myers, B. Neural regulation of the stress response: Glucocorticoid feedback mechanisms. Braz. J. Med. Biol. Res. Rev. Bras. Pesqui. Med. Biol. 2012, 45, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Palma-Gudiel, H.; Córdova-Palomera, A.; Leza, J.C.; Fañanás, L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neurosci. Biobehav. Rev. 2015, 55, 520–535. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Binder, E.B. Gene-environment interactions at the FKBP5 locus: Sensitive periods, mechanisms and pleiotropism. Genes Brain Behav. 2014, 13, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Numakawa, T.; Odaka, H.; Adachi, N. Actions of Brain-Derived Neurotrophic Factor and Glucocorticoid Stress in Neurogenesis. Int. J. Mol. Sci. 2017, 18, 2312. [Google Scholar] [CrossRef] [PubMed]
- Non, A.L.; Binder, A.M.; Barault, L.; Rancourt, R.C.; Kubzansky, L.D.; Michels, K.B. DNA methylation of stress-related genes and LINE-1 repetitive elements across the healthy human placenta. Placenta 2012, 33, 183–187. [Google Scholar] [CrossRef]
- Chen, J.; Li, Q.; Rialdi, A.; Mystal, E.; Ly, J.; Finik, J.; Davey, T.; Lambertini, L.; Nomura, Y. Influences of Maternal Stress during Pregnancy on the Epi/genome: Comparison of Placenta and Umbilical Cord Blood. J. Depress. Anxiety 2014, 3, 152. [Google Scholar] [CrossRef] [PubMed]
- Watkeys, O.J.; Kremerskothen, K.; Quidé, Y.; Fullerton, J.M.; Green, M.J. Glucocorticoid receptor gene (NR3C1) DNA methylation in association with trauma, psychopathology, transcript expression, or genotypic variation: A systematic review. Neurosci. Biobehav. Rev. 2018, 95, 85–122. [Google Scholar] [CrossRef]
- Labonté, B.; Azoulay, N.; Yerko, V.; Turecki, G.; Brunet, A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl. Psychiatry 2014, 4, e368. [Google Scholar] [CrossRef] [PubMed]
- McGowan, P.O.; Sasaki, A.; D’Alessio, A.C.; Dymov, S.; Labonté, B.; Szyf, M.; Turecki, G.; Meaney, M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009, 12, 342–348. [Google Scholar] [CrossRef]
- Perroud, N.; Rutembesa, E.; Paoloni-Giacobino, A.; Mutabaruka, J.; Mutesa, L.; Stenz, L.; Malafosse, A.; Karege, F. The Tutsi genocide and transgenerational transmission of maternal stress: Epigenetics and biology of the HPA axis. World J. Biol. Psychiatry 2014, 15, 334–345. [Google Scholar] [CrossRef] [PubMed]
- Vukojevic, V.; Kolassa, I.T.; Fastenrath, M.; Gschwind, L.; Spalek, K.; Milnik, A.; Heck, A.; Vogler, C.; Wilker, S.; Demougin, P.; et al. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. J. Neurosci. 2014, 34, 10274–10284. [Google Scholar] [CrossRef] [PubMed]
- Yehuda, R.; Flory, J.D.; Bierer, L.M.; Henn-Haase, C.; Lehrner, A.; Desarnaud, F.; Makotkine, I.; Daskalakis, N.P.; Marmar, C.R.; Meaney, M.J. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol. Psychiatry 2015, 77, 356–364. [Google Scholar] [CrossRef]
- Zannas, A.S.; Wiechmann, T.; Gassen, N.C.; Binder, E.B. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology 2016, 41, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Klengel, T.; Mehta, D.; Anacker, C.; Rex-Haffner, M.; Pruessner, J.C.; Pariante, C.M.; Pace, T.W.; Mercer, K.B.; Mayberg, H.S.; Bradley, B.; et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci. 2013, 16, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Thomassin, H.; Flavin, M.; Espinás, M.L.; Grange, T. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 2001, 20, 1974–1983. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Jia, M.; Hafner, K.; Baumert, J.; Wiechmann, T.; Pape, J.C.; Arloth, J.; Ködel, M.; Martinelli, S.; Roitman, M.; et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB-driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci. USA 2019, 116, 11370–11379. [Google Scholar] [CrossRef] [PubMed]
- Sluiter, F.; Incollingo Rodriguez, A.C.; Nephew, B.C.; Cali, R.; Murgatroyd, C.; Santos, H.P., Jr. Pregnancy associated epigenetic markers of inflammation predict depression and anxiety symptoms in response to discrimination. Neurobiol. Stress 2020, 13, 100273. [Google Scholar] [CrossRef]
- Monk, C.; Feng, T.; Lee, S.; Krupska, I.; Champagne, F.A.; Tycko, B. Distress During Pregnancy: Epigenetic Regulation of Placenta Glucocorticoid-Related Genes and Fetal Neurobehavior. Am. J. Psychiatry 2016, 173, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Walsh, K.; McCormack, C.A.; Webster, R.; Pinto, A.; Lee, S.; Feng, T.; Krakovsky, H.S.; O’Grady, S.M.; Tycko, B.; Champagne, F.A.; et al. Maternal prenatal stress phenotypes associate with fetal neurodevelopment and birth outcomes. Proc. Natl. Acad. Sci. USA 2019, 116, 23996–24005. [Google Scholar] [CrossRef]
- Heiss, J.A.; Just, A.C. Identifying mislabeled and contaminated DNA methylation microarray data: An extended quality control toolset with examples from GEO. Clin. Epigenetics 2018, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Perrier, F.; Novoloaca, A.; Ambatipudi, S.; Baglietto, L.; Ghantous, A.; Perduca, V.; Barrdahl, M.; Harlid, S.; Ong, K.K.; Cardona, A.; et al. Identifying and correcting epigenetics measurements for systematic sources of variation. Clin. Epigenetics 2018, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Saraei, S.; Suomi, T.; Kauko, O.; Elo, L.L.; Stegle, O. Phosphonormalizer: An R package for normalization of MS-based label-free phosphoproteomics. Bioinformatics 2018, 34, 693–694. [Google Scholar] [CrossRef] [PubMed]
- Salas, L.A.; Koestler, D.C.; Butler, R.A.; Hansen, H.M.; Wiencke, J.K.; Kelsey, K.T.; Christensen, B.C. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018, 19, 64. [Google Scholar] [CrossRef] [PubMed]
- Lam, L.L.; Emberly, E.; Fraser, H.B.; Neumann, S.M.; Chen, E.; Miller, G.E.; Kobor, M.S. Factors underlying variable DNA methylation in a human community cohort. Proc. Natl. Acad. Sci. USA 2012, 109 (Suppl. S2), 17253–17260. [Google Scholar] [CrossRef] [PubMed]
- Zilbauer, M.; Rayner, T.F.; Clark, C.; Coffey, A.J.; Joyce, C.J.; Palta, P.; Palotie, A.; Lyons, P.A.; Smith, K.G. Genome-wide methylation analyses of primary human leukocyte subsets identifies functionally important cell-type-specific hypomethylated regions. Blood 2013, 122, e52–e60. [Google Scholar] [CrossRef] [PubMed]
- Houseman, E.A.; Accomando, W.P.; Koestler, D.C.; Christensen, B.C.; Marsit, C.J.; Nelson, H.H.; Wiencke, J.K.; Kelsey, K.T. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012, 13, 86. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.C.; Yet, I.; Tsai, P.C.; Bell, J.T. coMET: Visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinform. 2015, 16, 131. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
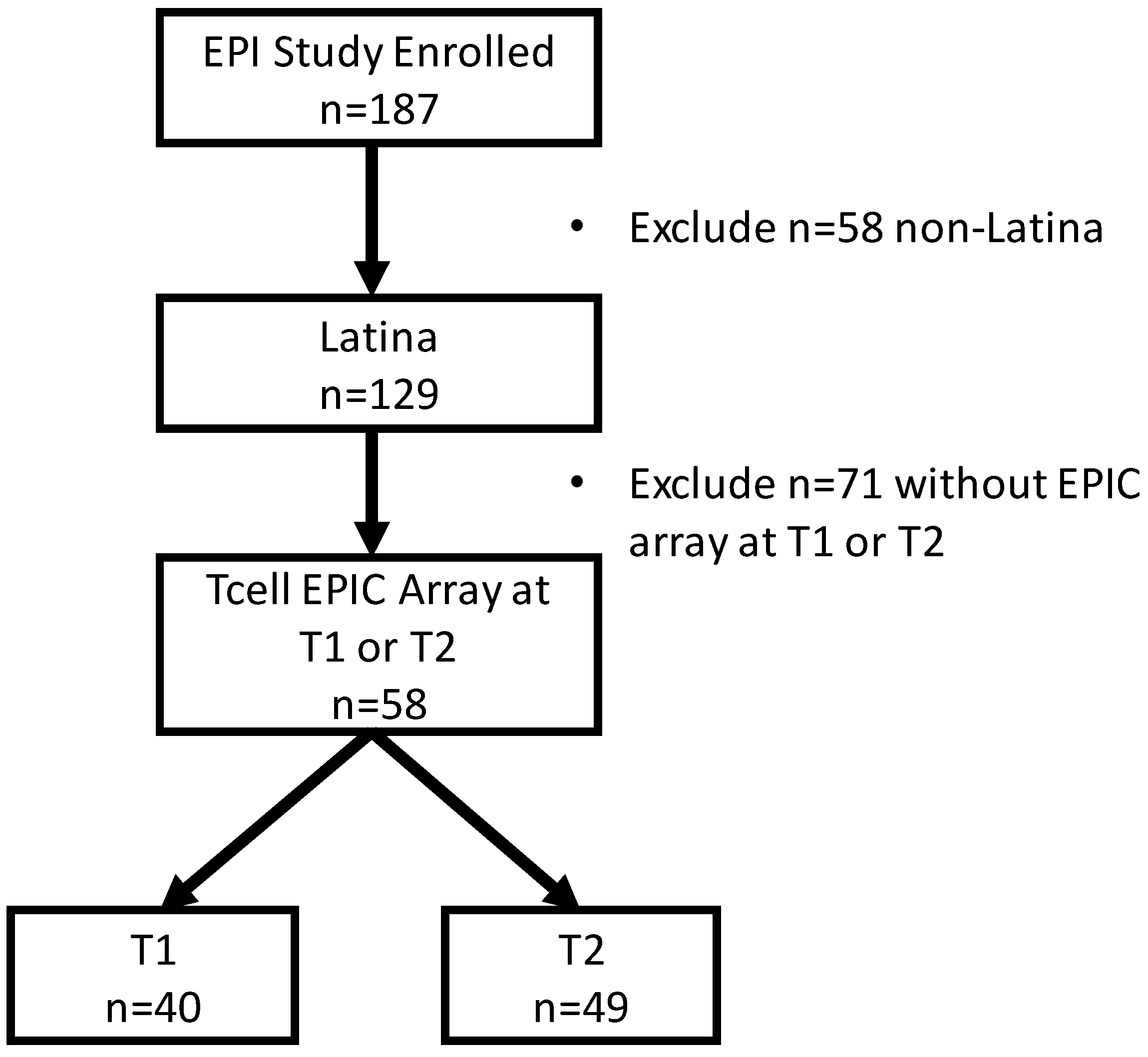
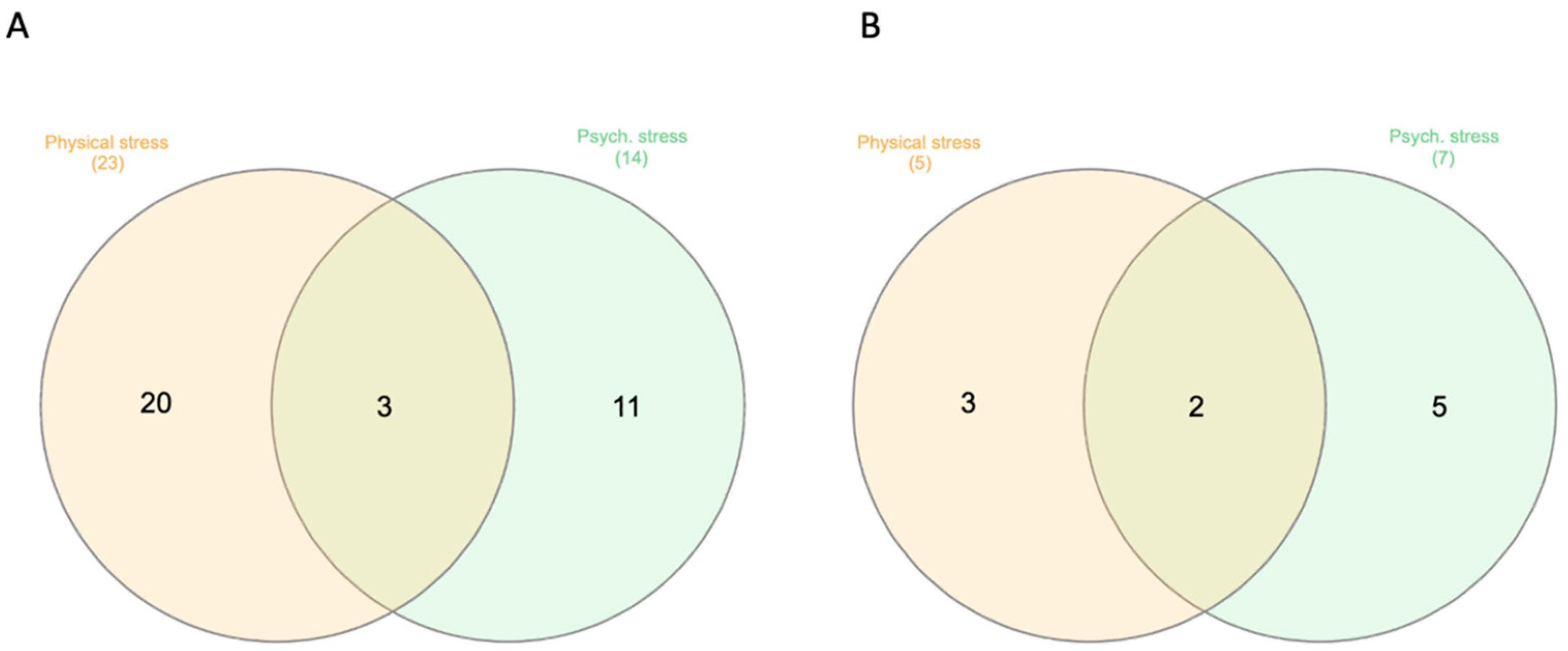
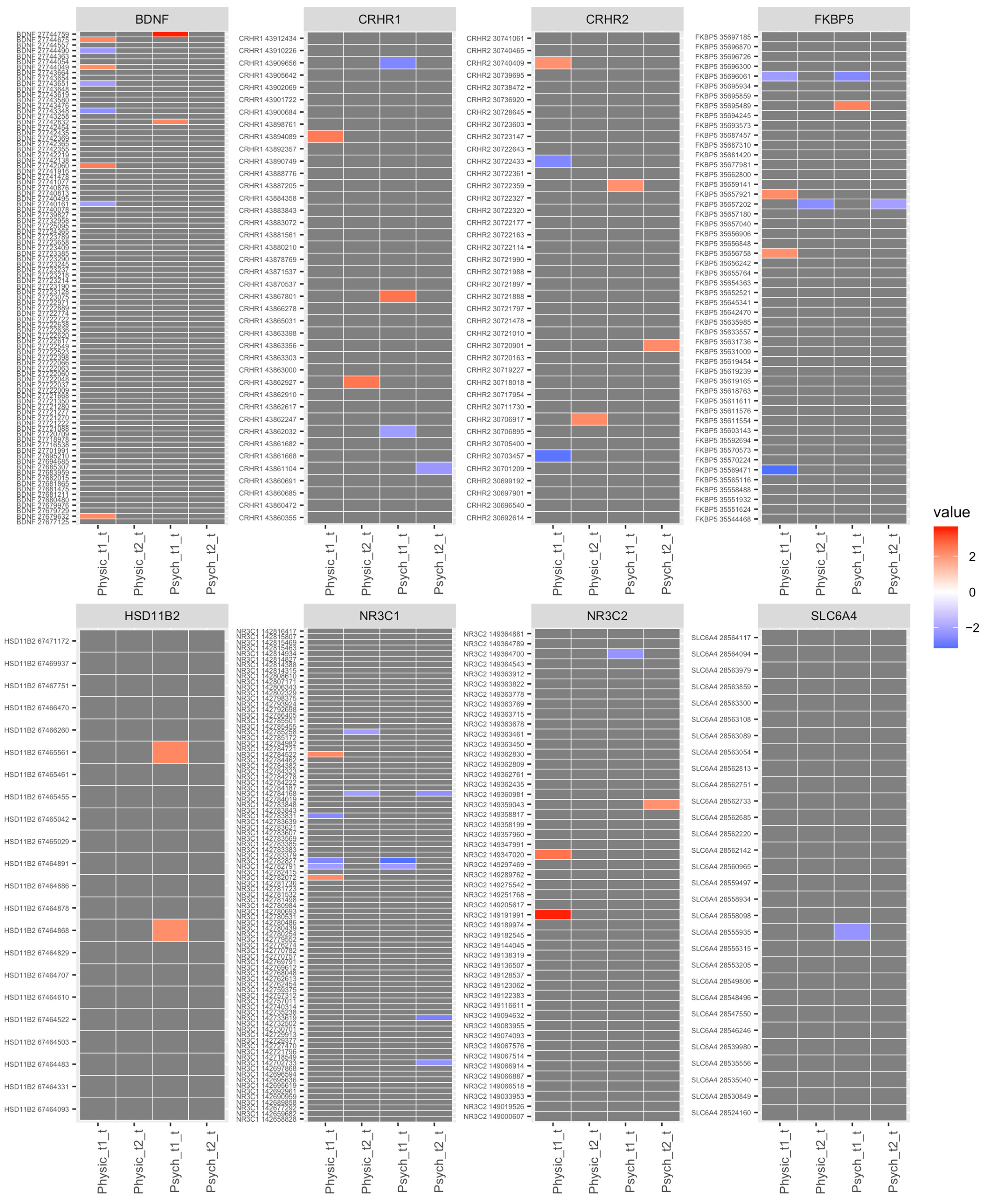
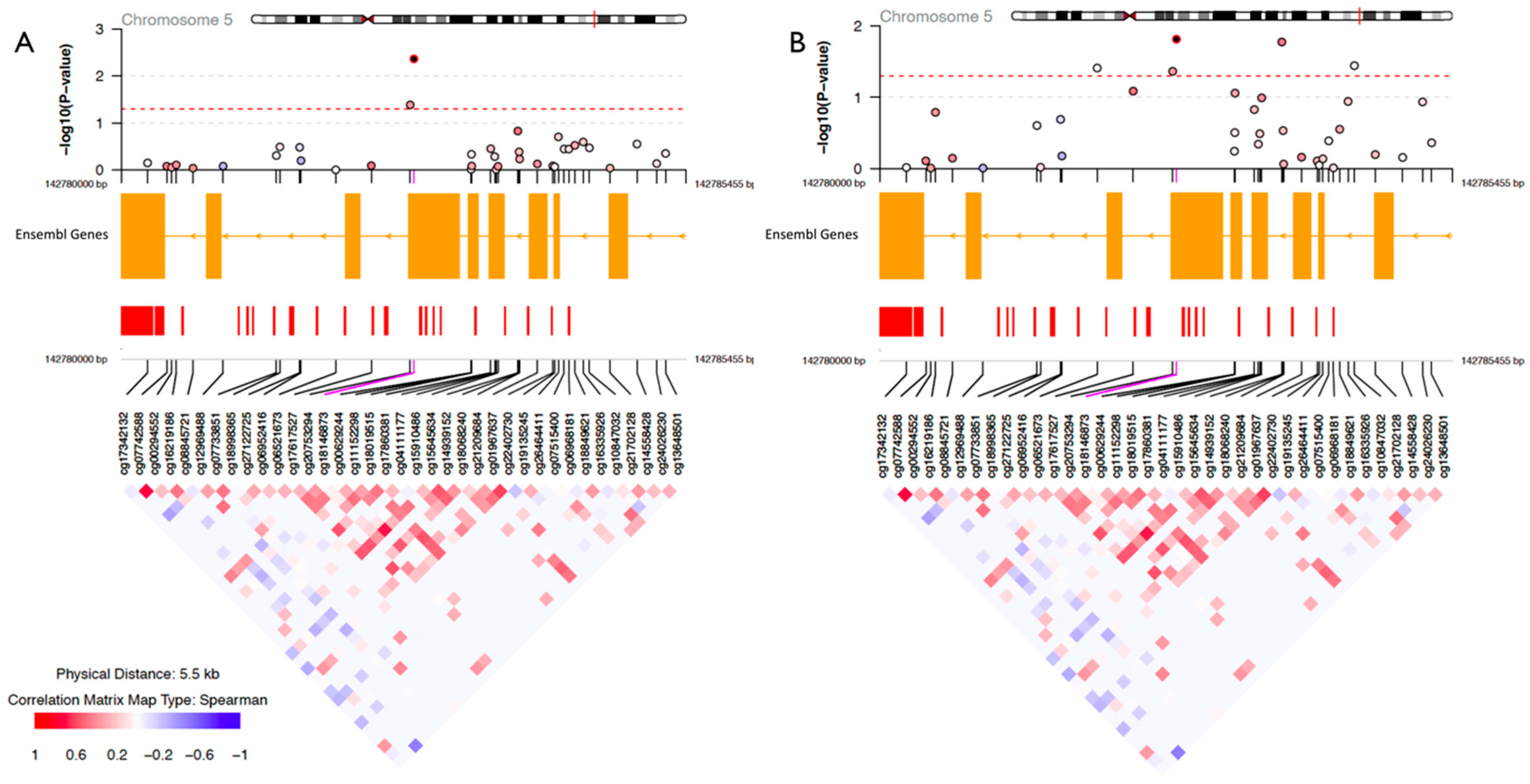
| Healthy (n = 34) | Psychologically Stressed (n = 15) | Physically Stressed (n = 9) | Total (N = 58) | |||||
|---|---|---|---|---|---|---|---|---|
| n (Mean) | % (s.d.) | n (Mean) | % (s.d.) | n (Mean) | % (s.d.) | n (Mean) | % (s.d.) | |
| Mean (s.d.) maternal age at enrollment (T1) | ||||||||
| 27.2 | 5.5 | 27.6 | 4.6 | 26.0 | 3.5 | 27.1 | 5.0 | |
| Race | ||||||||
| American Indian/Alaskan | 20 | 58.8 | 8 | 53.3 | 6 | 66.7 | 34 | 58.6 |
| Black/African American | 1 | 2.9 | 2 | 13.3 | 3 | 33.3 | 6 | 10.3 |
| White | 8 | 23.5 | 3 | 20.0 | 0 | 0.0 | 11 | 19.0 |
| Biracial | 2 | 5.9 | 0 | 0.0 | 0 | 0.0 | 2 | 3.4 |
| Other | 3 | 8.8 | 2 | 13.3 | 0 | 0.0 | 3 | 8.6 |
| Fetal sex | ||||||||
| Male | 21 | 61.8 | 6 | 40.0 | 1 | 11.1 | 28 | 48.3 |
| Female | 13 | 38.2 | 9 | 60.0 | 8 | 88.9 | 30 | 51.7 |
| Parity | ||||||||
| 0 | 30 | 88.2 | 12 | 80.0 | 8 | 88.9 | 50 | 86.2 |
| 1 | 4 | 11.8 | 3 | 20.0 | 1 | 11.1 | 8 | 13.8 |
| Gestational age at birth (weeks) | 39.47 | 1.78 | 38.8 | 2.7 | 39.1 | 1.47 | 39.24 | 2.0 |
| Highest education completed | ||||||||
| High School graduate/GED | 22 | 64.7 | 10 | 71.4 | 8 | 88.9 | 40 | 70.2 |
| Associate degree/College graduate or higher | 12 | 35.3 | 4 | 28.6 | 1 | 11.1 | 17 | 29.8 |
| Annual household income | ||||||||
| ≤$15,000 | 6 | 17.6 | 5 | 33.3 | 1 | 11.1 | 12 | 20.7 |
| >$15,000–$50,000 | 14 | 41.2 | 7 | 46.7 | 7 | 77.8 | 28 | 48.3 |
| ≥$50,000 | 14 | 41.2 | 3 | 20.0 | 1 | 11.1 | 18 | 31.0 |
| Medicaid | ||||||||
| Yes | 21 | 61.8 | 12 | 85.7 | 8.0 | 88.9 | 41 | 71.9 |
| No | 13 | 38.2 | 2 | 14.3 | 1.0 | 11.1 | 16 | 28.1 |
| Current smoker | ||||||||
| Yes | 0 | 0 | 0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| No | 34 | 100 | 14 | 100.0 | 9.0 | 100.0 | 57 | 100.0 |
| Ever smoker | ||||||||
| Yes | 9 | 26.5 | 6 | 42.9 | 4 | 44.4 | 19 | 33.3 |
| No | 25 | 73.5 | 8 | 57.1 | 5 | 55.6 | 38 | 66.7 |
| Gene | Number of CpGs |
|---|---|
| BDNF | 90 |
| CRHR1 | 40 |
| CRHR2 | 40 |
| FKBP5 | 50 |
| HSD11B2 | 22 |
| NR3C1 | 88 |
| NR3C2 | 49 |
| SLC6A4 | 30 |
| Total | 409 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barcelona, V.; Abuaish, S.; Lee, S.; Harkins, S.; Butler, A.; Tycko, B.; Baccarelli, A.A.; Walsh, K.; Monk, C.E. Stress and DNA Methylation of Blood Leukocytes among Pregnant Latina Women. Epigenomes 2023, 7, 27. https://doi.org/10.3390/epigenomes7040027
Barcelona V, Abuaish S, Lee S, Harkins S, Butler A, Tycko B, Baccarelli AA, Walsh K, Monk CE. Stress and DNA Methylation of Blood Leukocytes among Pregnant Latina Women. Epigenomes. 2023; 7(4):27. https://doi.org/10.3390/epigenomes7040027
Chicago/Turabian StyleBarcelona, Veronica, Sameera Abuaish, Seonjoo Lee, Sarah Harkins, Ashlie Butler, Benjamin Tycko, Andrea A. Baccarelli, Kate Walsh, and Catherine E. Monk. 2023. "Stress and DNA Methylation of Blood Leukocytes among Pregnant Latina Women" Epigenomes 7, no. 4: 27. https://doi.org/10.3390/epigenomes7040027
APA StyleBarcelona, V., Abuaish, S., Lee, S., Harkins, S., Butler, A., Tycko, B., Baccarelli, A. A., Walsh, K., & Monk, C. E. (2023). Stress and DNA Methylation of Blood Leukocytes among Pregnant Latina Women. Epigenomes, 7(4), 27. https://doi.org/10.3390/epigenomes7040027







