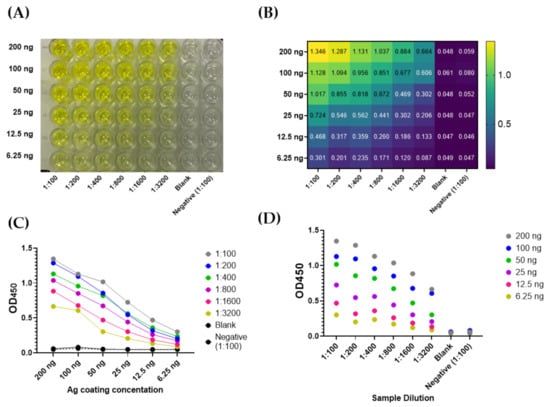
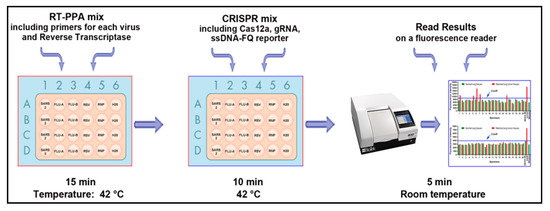
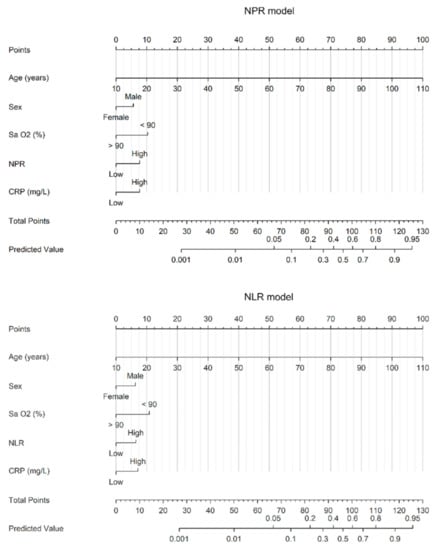
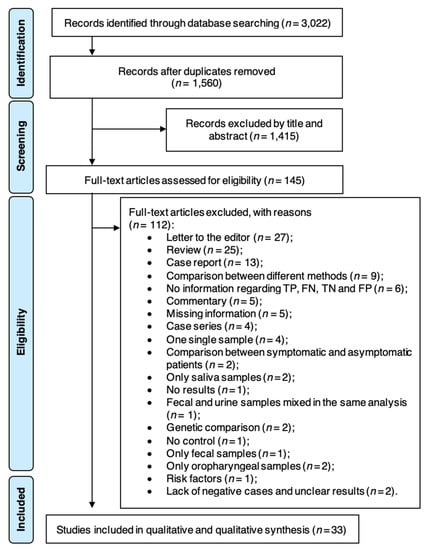
Diagnostic Virology
A topical collection in Diagnostics (ISSN 2075-4418). This collection belongs to the section "Diagnostic Microbiology and Infectious Disease".
Viewed by 70137Editor
Interests: HIV; antivirals and vaccines; drug resistance; pathogenesis; antibody neutralization; virus evolution
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
In these troubled COVID-19 times we are living in, it has become more evident than ever that rapid, deployable, and highly sensitive, specific and precise diagnostic assays can save many lives. Together with the established qPCR-based and isothermal amplification-based methods, new technologies such as those based on CRISPR-Cas13 system and metagenomics are emerging and are enabling the rapid and sensitive detection and identification of multiple viruses at the same time with an unprecedented quality, both in the lab and outside the lab (PoC assays). These molecular methods of diagnostics rely on nucleic acid extraction methods whose performance characteristics may be very diverse and need a better assessment. High quality serologic assays are also paramount for the diagnosis and management of virus diseases, and for monitoring the response to vaccination. A better understanding of the advantages and limitations of currently available molecular and serologic assays for the diagnosis, treatment management, and cure assessment of each virus disease is needed. There is no better time for this Topical Collection in diagnostic virology. I would like to invite you to submit your best review or original work on all aspects of diagnostic virology mentioned above and others that I have missed to this topical collection issue. We are particularly seeking top level contributions on the development, validation, and implementation of molecular and serologic assays for the diagnosis, prognosis, and management of diseases caused by human viruses in all settings from the lab to the house and the bed of the patient.
Prof. Dr. Nuno Taveira
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Diagnostics is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2600 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Detecton and identification of viral pathogens
- Development and validation of diagnostic tests (molecular, antigenic and serologic)
- Implementation of diagnostic tests in different settings and geographies
- Performance of nucleic acid extraction tests
- Point of care assays
- Multiplex assays
- CRISPR diagnostics
- Metagenomic diagnostics
- Drug resistance assays
- Diagnostic virology and public health