- Review
Single-Cell Transcriptomics and Computational Frameworks for Target Discovery in Cancer
- Martina Tarozzi,
- Nicolas Riccardo Derus and
- Gastone Castellani
- + 2 authors
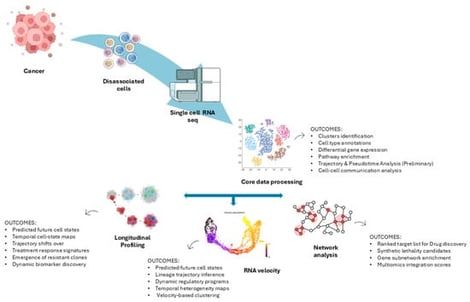
Single-cell transcriptomics has redefined our understanding of cancer by exposing the complexity of tumor ecosystems and their therapeutic vulnerabilities. scRNA-seq studies have identified lineage hierarchies, immune evasion programs, and resistance-associated states across solid and liquid tumors, informing biomarker development and drug discovery. Advanced computational frameworks integrate these data with longitudinal profiling, RNA velocity, and network diffusion to prioritize targets and predict therapeutic response. Emerging multi-omics approaches further expand the scope of precision oncology by linking genetic alterations, protein-level markers, and spatial context to functional states. This narrative review aims to synthesize current applications of single-cell transcriptomics for target discovery, highlight computational frameworks that translate high-dimensional data into actionable insights, and explore how multi-omics integration is shaping future directions. By bridging molecular complexity with target prioritization, these approaches hold promise for translating single-cell insights into clinically actionable biomarkers and therapeutic strategies for personalized cancer treatment and rational drug development.
3 February 2026