- Communication
A Proteomic View of Butterfly Metamorphosis
- Andrew Hesketh,
- Juned Kadiwala and
- Vaishnavi Ravikumar
- + 4 authors
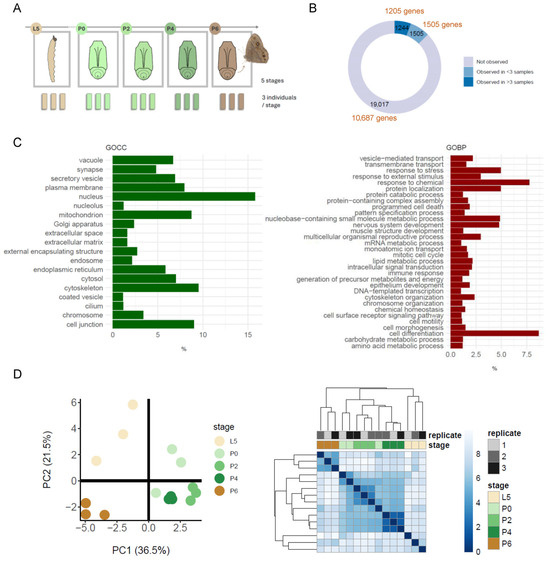
Background: Insect metamorphosis is one of the most fascinating developmental processes in the natural world. Complete metamorphosis requires the breakdown and reorganisation of larval tissues and the coordinated construction and development of adult structures. The molecular events that achieve this transformation are, however, incompletely understood, and there is a particular shortage of data describing changes in protein abundance that occur during the process. Methods: Here, using a label-free quantitative bottom-up approach, we perform a novel whole-organism proteomic analysis of consecutive developmental stages of male Bicyclus anynana butterflies as they develop from caterpillars into adults via pupation. Results: Our analysis generated a dynamic reference dataset representing 2749 detected proteins. Statistical analysis identified 90 proteins changing significantly in abundance during metamorphosis, and functional interpretation highlights cuticle formation, apoptosis and autophagy during the pupal stages, and the up-regulation of respiration and energy metabolism upon completion of the fully formed adult. A preliminary search for potential peptide phosphorylation modifications identified 15 candidates, including three proteins with roles in muscle function. Conclusions: The study provides a basis for future protein-level analysis of butterfly metamorphosis and suggests the importance of dissecting the post-translational regulation associated with this fascinating developmental transformation.
18 December 2025