- Article
Xenograft-Derived Human Breast Cancer Organoids Can Form Chimeras with Host Mouse Mammary Epithelial Cells Which Promote Tumor Cell Proliferation
- Hiroyuki Uematsu,
- Chieko Saito and
- Masahiro Inoue
- + 8 authors
Breast cancer progression and treatment responsiveness are significantly influenced by the tumor microenvironment. Therefore, transplantation into the mammary fat pad is widely employed to establish a mouse xenograft model of breast cancer. This study reports chimeric organoids derived from breast cancer xenografts composed of human and mouse cells. During passaging of an organoid line derived from breast cancer xenografts, characteristic cell clusters composed of smaller cells were observed. Immunostaining with a mouse-specific antibody revealed that the smaller cells were mouse cells composed of luminal- and basal-like cells. Chimeric organoids were observed in four of the six xenograft-derived organoid lines. Organoids composed solely of human cells rapidly diminished after passaging, with chimeric and mouse-cell-only organoids becoming predominant. When human breast cancer cells were co-cultured with mouse mammary epithelial cells, chimeras were frequently observed. The PCNA positivity rate in breast cancer cells within chimeras was higher than that in breast cancer cells within organoids composed solely of human cells. These findings indicate that xenograft-derived breast cancer organoids frequently contain mouse cells and that mouse mammary epithelial cells promote the proliferation of human breast cancer cells.
6 February 2026



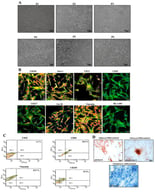
![The presence of mouse cells in organoids derived from xenografts. (A) During the passaging process of M63 breast cancer organoids, cell clusters of differing sizes were observed. The upper panels show large cell clusters, while the lower panels show small cell clusters. Left panels: bright-field images; right panels: Hoechst 33342 staining. Scale bar: 20 μm. (B) Microscopic and immunofluorescence (IF) staining images of spheroids derived from large (upper panels) and small (lower panels) cell clusters. mCyclophilin A (green): staining with a mouse-specific anti-cyclophilin A antibody; 4′,6-diamidino-2-phenylindole (DAPI, blue). Scale bars: 100 μm (brightfield), 50 μm (hematoxylin and eosin [HE], mCyclophilin A and DAPI). (C) IF staining of serial sections of spheroids derived from mouse cells isolated from M63 breast cancer organoids. Nuclei were counterstained with DAPI (blue). Scale bar, 50 μm. (D) Microscopic and IF staining images of mouse mammary organoids. IF images were obtained from serial sections. Nuclei were counterstained with DAPI (blue). Scale bar: 100 μm (brightfield), 50 μm (IF staining).](https://mdpi-res.com/cdn-cgi/image/w=470,h=317/https://mdpi-res.com/organoids/organoids-05-00006/article_deploy/html/images/organoids-05-00006-g001-550.jpg)