Organoids as Next-Generation Models for Tumor Heterogeneity, Personalized Therapy, and Cancer Research: Advancements, Applications, and Future Directions
Abstract
1. Introduction
1.1. Cancer Complexity and Model Limitations
1.2. Rise of Organoid Systems
1.3. Integration into Cancer Research
1.4. Structure and Objectives of the Review
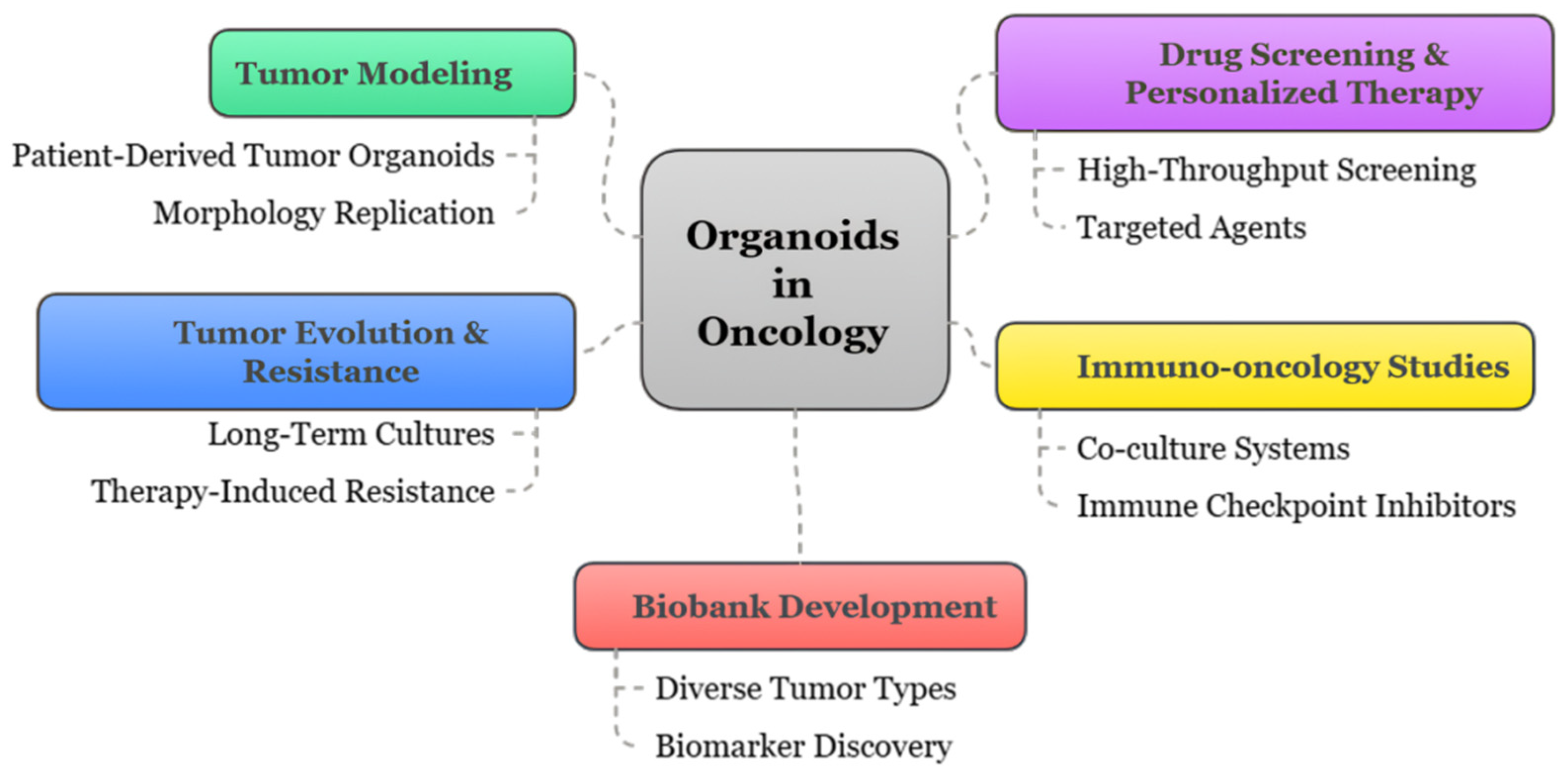
2. Organoids: Concept and Technology
2.1. Definition and Biological Basis
2.1.1. What Defines an Organoid
2.1.2. Morphology, Self-Organization, and Differentiation
2.1.3. Organoid Fidelity in Cancer Modeling
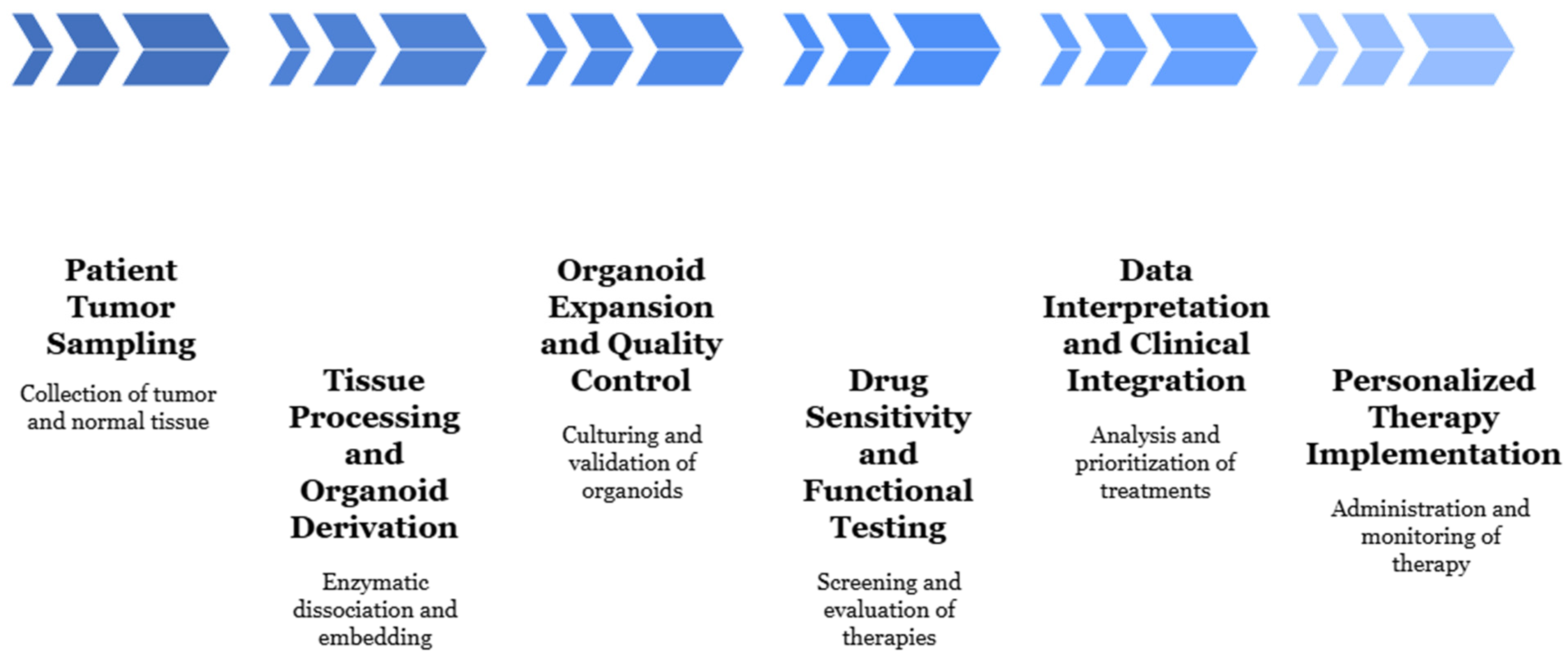
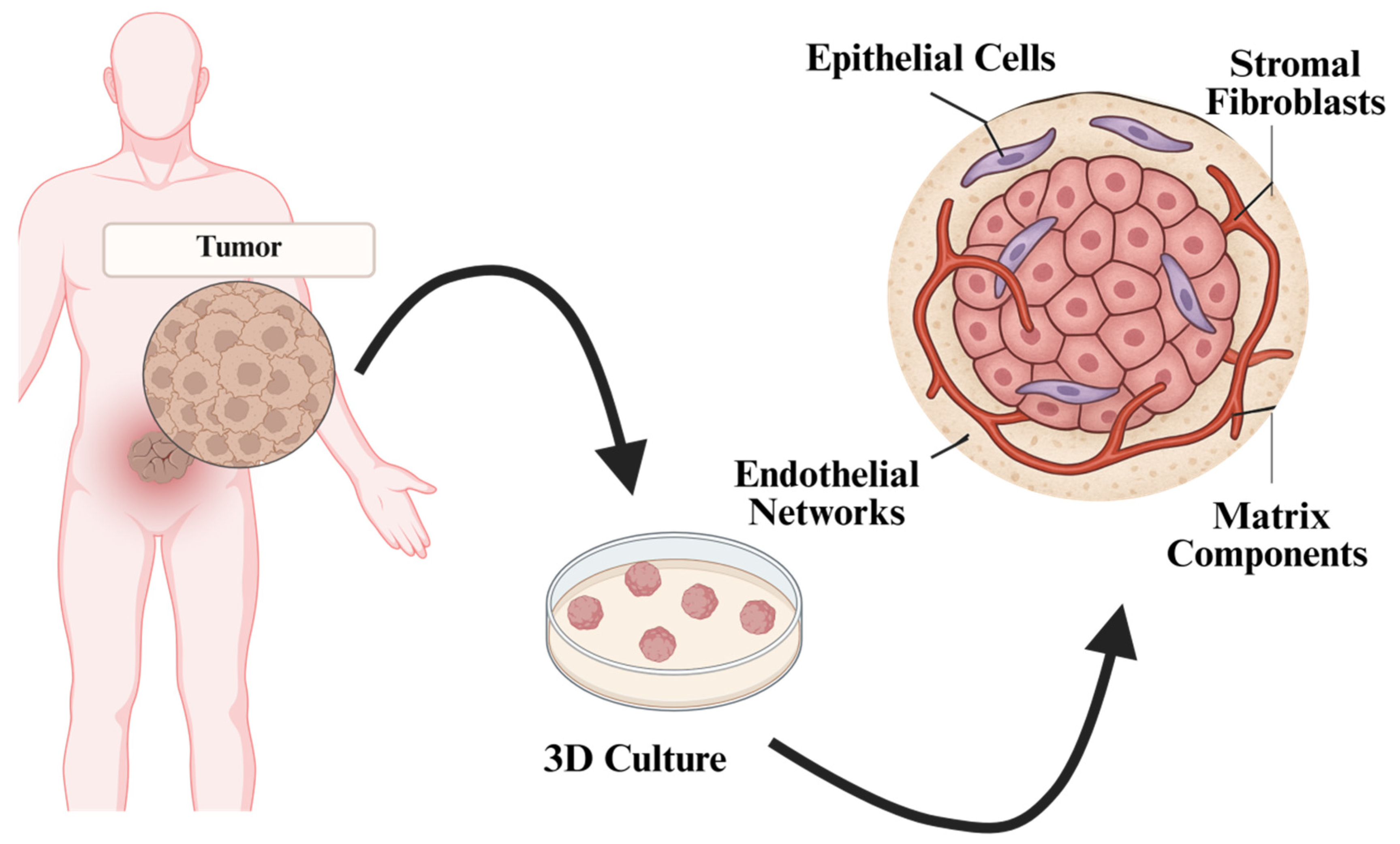
2.2. Techniques for Organoid Development in the Context of Tumor Modeling
2.2.1. Source Cell Types
Adult Stem Cells (ASCs) from Tumor Tissues
Cancer Stem Cells (CSCs)
Induced Pluripotent Stem Cells (iPSCs)
Embryonic Stem Cells (ESCs)
Tissue-Resident Progenitor Cells
Mesenchymal Stem Cells (MSCs)
Circulating Tumor Cells (CTCs)
Engineered Synthetic Progenitors
2.2.2. Culture Systems
Scaffold-Based Culture Systems
Scaffold-Free Culture Systems
2.2.3. Protocols and Maintenance
Media Components and Niche Factors
Growth Timeline and Batch Scaling
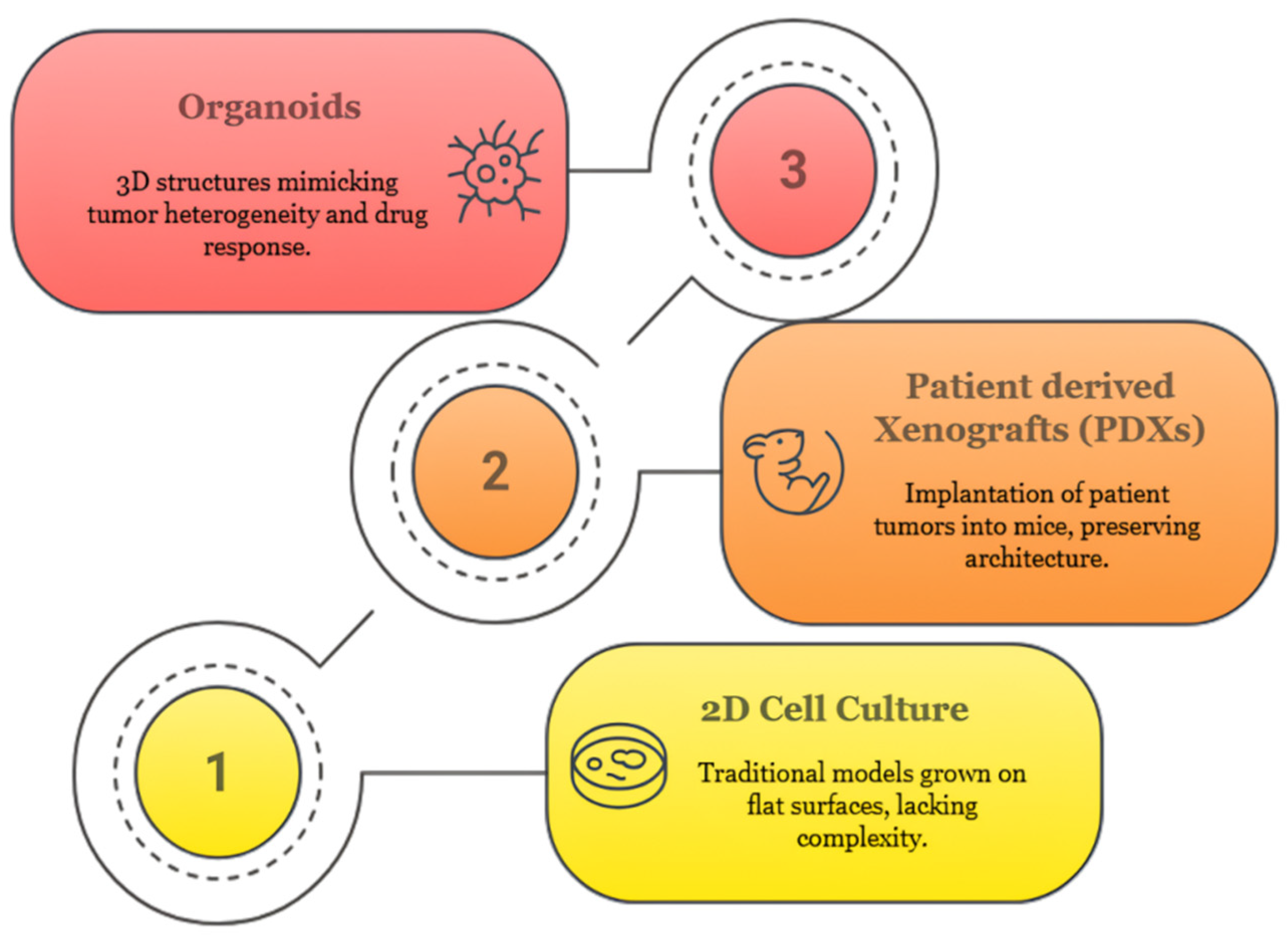
2.3. Comparison with Other Preclinical Models
2.3.1. Two-Dimensional (2D) Cell Lines
2.3.2. Three-Dimensional (3D) Spheroids
2.3.3. Patient-Derived Xenografts (PDXs)
2.3.4. Organoids: A New Paradigm in Preclinical Modeling
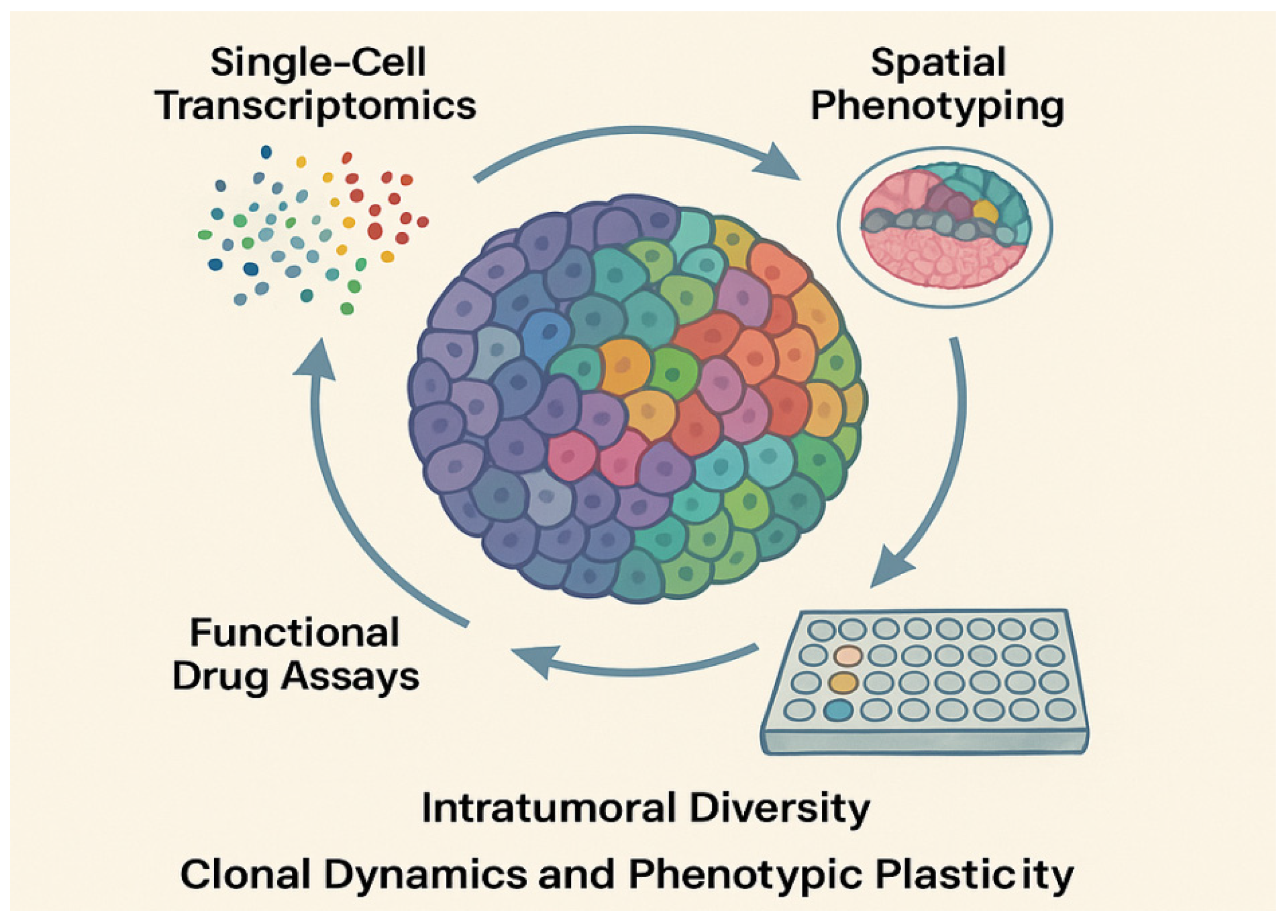
3. Tumor Heterogeneity and Organoids
3.1. Dimensions of Tumor Heterogeneity
3.1.1. Genetic and Epigenetic Variability
3.1.2. Spatial and Temporal Heterogeneity
3.1.3. Functional Heterogeneity in Drug Resistance
3.2. Organoids as Models for Tumor Heterogeneity
3.2.1. Preservation of Tumor Architecture and Clonal Diversity
3.2.2. Clonal Evolution and Single-Cell Resolution Studies
3.2.3. Subtype Stratification and Plasticity Assessment
3.3. Patient-Derived Organoid Biobanks
3.3.1. International Consortia and Standardization Efforts
3.3.2. Cancer Type Representation and Sample Diversity
3.3.3. Integration with Clinical Data and Predictive Platforms
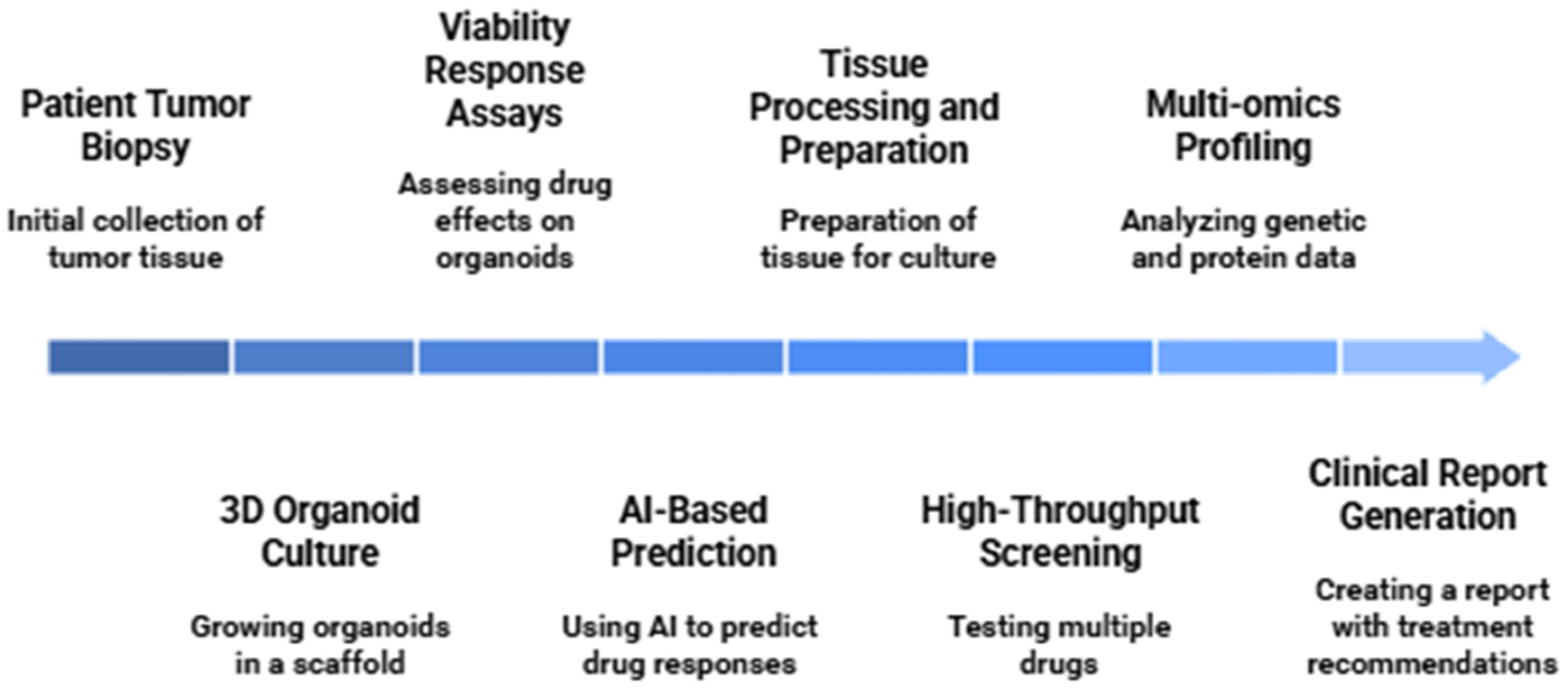
4. Organoids in Personalized Cancer Therapy
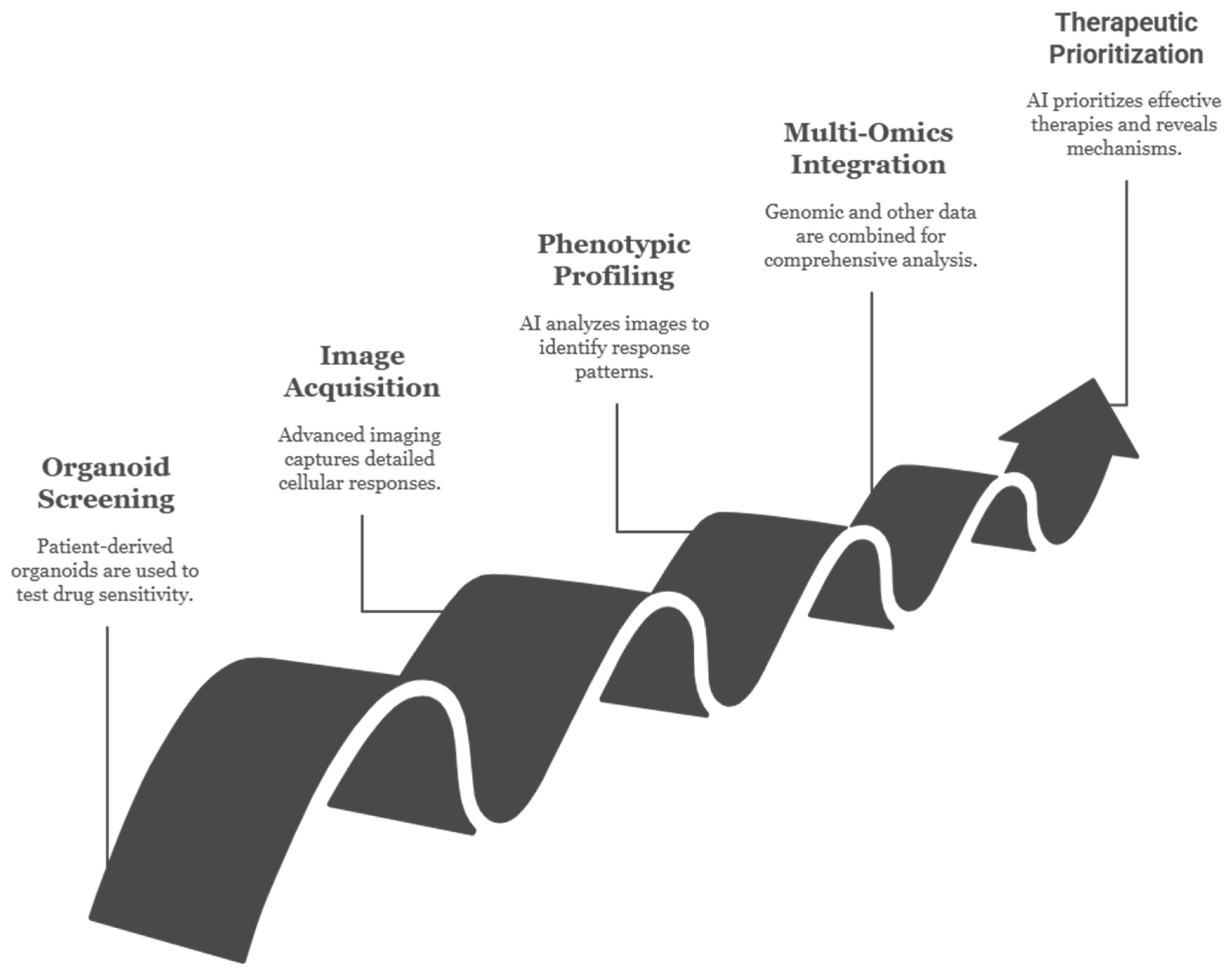
4.1. Drug Screening and Predictive Testing
4.1.1. High-Throughput Pharmacotyping Platforms
4.1.2. Organoid–Drug Response Correlation with Clinical Data
4.1.3. Resistance Profiling and Secondary Screening
4.2. Immune-Oncology Applications
4.2.1. Organoid–Immune Cell Co-Culture
4.2.2. Immunotherapy Biomarker Testing
4.2.3. Checkpoint Inhibitors and CAR-T Cell Testing in PDOs
4.2.4. Future Directions
- Emerging innovations continue to enhance the predictive and translational potential of PDO–immune co-culture platforms.
- High-content imaging and AI integration are enabling real-time tracking of immune–tumor interactions, immune synapse dynamics, and cytokine signaling within live organoids, facilitating quantitative immune response profiling [161].
- Complex tumor microenvironment (TME) modeling through the addition of stromal fibroblasts, endothelial cells, and extracellular matrix components is refining the physiological relevance of organoid-based immune assays [162].
- Mathematical modeling and systems immunology approaches are being employed to simulate CAR-T dynamics, antigen heterogeneity, and immunosuppressive gradients, guiding rational design of immunotherapies and dosing strategies [163].
- Clinical trial integration of PDO-based immune stratification tools is underway, with organoid response data being incorporated into biomarker-driven, umbrella, and basket trial designs.
4.3. Clinical Case Studies: Real-World Translation of PDO-Guided Therapy
4.3.1. Predicting FOLFOX Response in Colorectal Cancer: A Landmark Prospective Study
4.3.2. Cetuximab Efficacy in RAS-Wild-Type CRC: Beyond Genomic Predictors
4.3.3. PARP Inhibitor Sensitivity in BRCA-Deficient and BRCA-Wild-Type TNBC
4.3.4. Dissecting EGFR-TKI Sensitivity in NSCLC: From Canonical to Atypical EGFR Mutations
4.3.5. Functional TKI Sensitivity in EGFR-Wild-Type NSCLC Revealed by PDO Profiling
4.4. Real-Time Therapy Guidance Using PDOs
5. Fundamental and Translational Applications
5.1. Modeling Cancer Initiation and Progression
5.1.1. Gene Editing (CRISPR/Cas9) in Organoids
5.1.2. APC, TP53, and KRAS Mutations in CRC Modeling
5.1.3. Phenotype Tracking Through Lineage Tracing
5.2. Microbiome–Tumor–Host Axis
5.2.1. GI Organoids and Microbiota Co-Culture
5.2.2. Role of Short-Chain Fatty Acids and Pathogens
5.3. Tumor Microenvironment Integration
5.3.1. Fibroblasts, Endothelial Cells, ECM Modeling
5.3.2. Paracrine Signaling and Angiogenesis Assays
5.4. Organoids in Cancer Metabolism Studies
5.4.1. Modeling Metabolic Reprogramming
5.4.2. Metabolic Dependencies and Therapy Resistance
5.4.3. Hypoxia and Microenvironmental Influence
5.4.4. Epigenetic Regulation and Plasticity
5.4.5. Translational Perspectives
6. Challenges and Limitations
6.1. Standardization Issues
6.1.1. Inter-Laboratory Variability
6.1.2. Matrix and Media Inconsistencies
6.2. Incomplete Physiological Representation
6.2.1. Absence of Vascularization
6.2.2. Limited Neural and Hormonal Inputs
6.2.3. Incomplete Immune Representation
6.3. Scalability, Cost, and Clinical Translation
6.3.1. Limited Scalability and GMP Compliance
6.3.2. High Operational Costs
6.4. Regulatory and Ethical Considerations
6.4.1. Lack of Regulatory Frameworks
6.4.2. Ethical and Data Governance Issues
7. Future Perspectives and Innovations
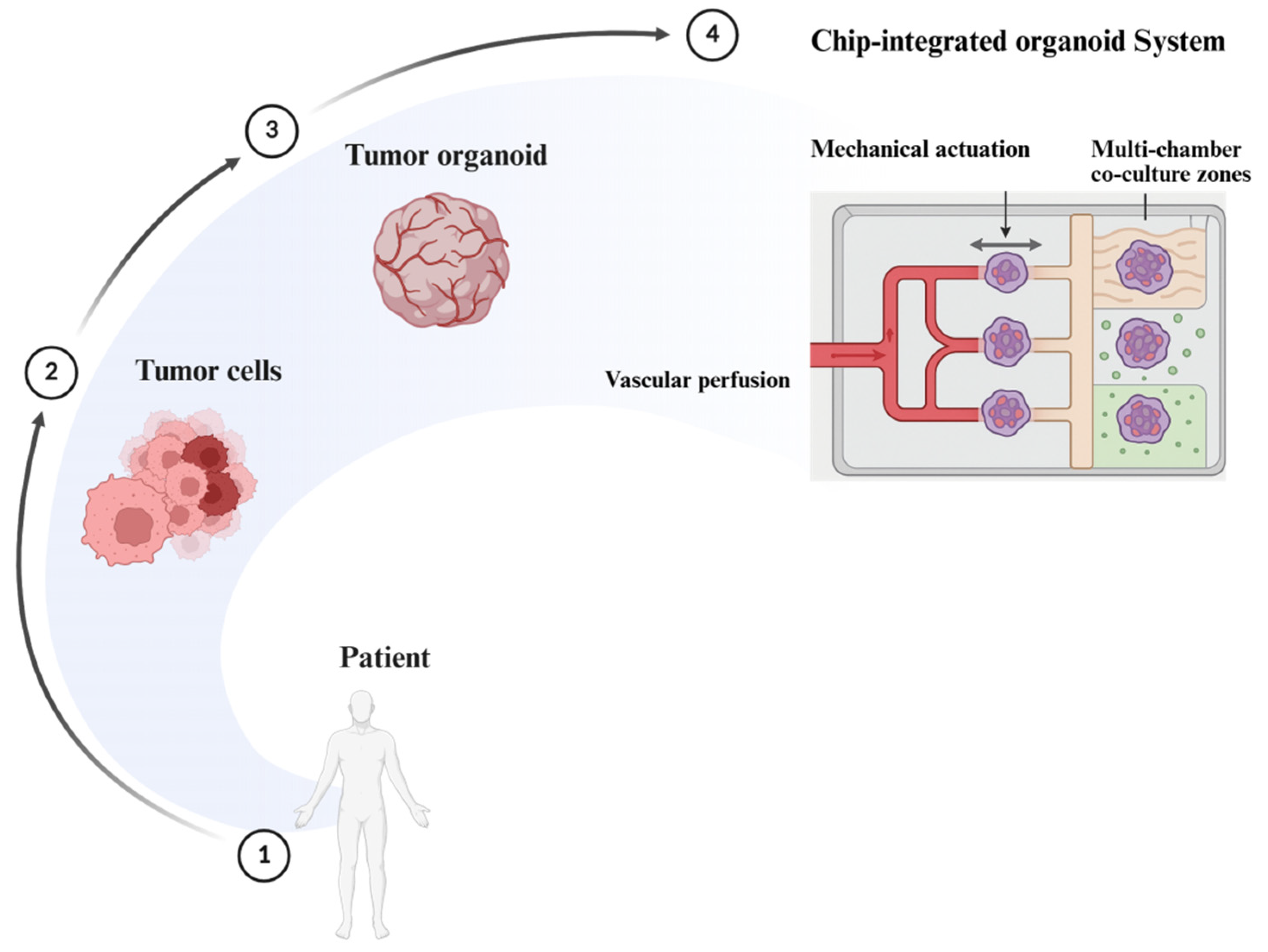
7.1. Organoid-on-a-Chip
7.1.1. Microfluidic Perfusion Systems
7.1.2. Spatiotemporal Simulation of Organ-Level Cues
7.2. Organoid Libraries and CRISPR Screens
7.2.1. Isogenic Organoid Banks
7.2.2. Functional Genomic Screens (Synthetic Lethality)
7.3. Organoid Image-to-Response Prediction
7.4. Strategic Gaps and Translational Priorities in Organoid Oncology
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biswas, A.; De, S. Drivers of dynamic intratumor heterogeneity and phenotypic plasticity. Am. J. Physiol. Cell Physiol. 2021, 320, C750–C760. [Google Scholar] [CrossRef]
- Zhang, A.; Miao, K.; Sun, H.; Deng, C.X. Tumor heterogeneity reshapes the tumor microenvironment to influence drug resistance. Int. J. Biol. Sci. 2022, 18, 3019. [Google Scholar] [CrossRef]
- Sajjad, H.; Imtiaz, S.; Noor, T.; Siddiqui, Y.H.; Sajjad, A.; Zia, M. Cancer models in preclinical research: A chronicle review of advancement in effective cancer research. Anim. Model. Exp. Med. 2021, 4, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Habanjar, O.; Diab-Assaf, M.; Caldefie-Chezet, F.; Delort, L. 3D cell culture systems: Tumor application, advantages, and disadvantages. Int. J. Mol. Sci. 2021, 22, 12200. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, W.; Cai, C.; Zhang, H.; Shen, H.; Han, Y. Patient-derived xenograft models in cancer therapy: Technologies and applications. Signal Transduct. Target. Ther. 2023, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Cai, C.; Deng, W.; Shi, Y.; Li, L.; Wang, C.; Zhang, J.; Rong, M.; Liu, J.; Fang, B.; et al. Landscape of human organoids: Ideal model in clinics and research. Innov. 2024, 5, 100620. [Google Scholar] [CrossRef]
- Shao, Y.; Fu, J. Engineering multiscale structural orders for high-fidelity embryoids and organoids. Cell Stem Cell 2022, 29, 722–743. [Google Scholar] [CrossRef]
- Furbo, S.; Urbano, P.C.; Raskov, H.H.; Troelsen, J.T.; Kanstrup Fiehn, A.M.; Gögenur, I. Use of patient-derived organoids as a treatment selection model for colorectal cancer: A narrative review. Cancers 2022, 14, 1069. [Google Scholar] [CrossRef]
- Qu, S.; Xu, R.; Yi, G.; Li, Z.; Zhang, H.; Qi, S.; Huang, G. Patient-derived organoids in human cancer: A platform for fundamental research and precision medicine. Mol. Biomed. 2024, 5, 6. [Google Scholar] [CrossRef]
- Xian, L.; Zhao, P.; Chen, X.; Wei, Z.; Ji, H.; Zhao, J.; Liu, W.; Li, Z.; Liu, D.; Han, X.; et al. Heterogeneity, inherent and acquired drug resistance in patient-derived organoid models of primary liver cancer. Cell. Oncol. 2022, 45, 1019–1036. [Google Scholar] [CrossRef]
- Singh, D.; Thakur, A.; Rakesh kumar, A. Advancements in Organoid-Based Drug Discovery: Revolutionizing Precision Medicine and Pharmacology. Drug Dev. Res. 2025, 86, e70121. [Google Scholar] [CrossRef] [PubMed]
- Bolck, H.A.; Corrò, C.; Kahraman, A.; von Teichman, A.; Toussaint, N.C.; Kuipers, J.; Chiovaro, F.; Koelzer, V.H.; Pauli, C.; Moritz, W.; et al. Tracing clonal dynamics reveals that two-and three-dimensional patient-derived cell models capture tumor heterogeneity of clear cell renal cell carcinoma. Eur. Urol. Focus 2021, 7, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Arana, Á.J.; González-Llera, L.; Barreiro-Iglesias, A.; Sánchez, L. Emerging Frontiers in Zebrafish Embryonic and Adult-Derived Cell Lines. Int. J. Mol. Sci. 2025, 26, 4351. [Google Scholar] [CrossRef] [PubMed]
- Serelli-Lee, V.; Ito, K.; Koibuchi, A.; Tanigawa, T.; Ueno, T.; Matsushima, N.; Imai, Y. A state-of-the-art roadmap for biomarker-driven drug development in the era of personalized therapies. J. Pers. Med. 2022, 12, 669. [Google Scholar] [CrossRef]
- Brassard, J.A.; Nikolaev, M.; Hübscher, T.; Hofer, M.; Lutolf, M.P. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 2021, 20, 22–29. [Google Scholar] [CrossRef]
- Spagnol, G.; Sensi, F.; De Tommasi, O.; Marchetti, M.; Bonaldo, G.; Xhindoli, L.; Noventa, M.; Agostini, M.; Tozzi, R.; Saccardi, C. Patient derived organoids (PDOs), extracellular matrix (ECM), tumor microenvironment (TME) and drug screening: State of the art and clinical implications of ovarian cancer organoids in the era of precision medicine. Cancers 2023, 15, 2059. [Google Scholar] [CrossRef]
- Nikokiraki, C.; Psaraki, A.; Roubelakis, M.G. The potential clinical use of stem/progenitor cells and organoids in liver diseases. Cells 2022, 11, 1410. [Google Scholar] [CrossRef]
- Elmentaite, R.; Domínguez Conde, C.; Yang, L.; Teichmann, S.A. Single-cell atlases: Shared and tissue-specific cell types across human organs. Nat. Rev. Genet. 2022, 23, 395–410. [Google Scholar] [CrossRef]
- Kang, D.H.; Lee, J.; Im, S.; Chung, C. Navigating the complexity of resistance in lung cancer therapy: Mechanisms, organoid models, and strategies for overcoming treatment failure. Cancers 2024, 16, 3996. [Google Scholar] [CrossRef]
- Ng, A.S.; Chan, D.K. Commonalities and differences in the mutational signature and somatic driver mutation landscape across solid and hollow viscus organs. Oncogene 2023, 42, 2713–2724. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Alam, K.; Roy, N.S.; Kaur, K.; Kaity, S.; Ravichandiran, V.; Roy, S. Exploring the interaction between extracellular matrix components in a 3D organoid disease model to replicate the pathophysiology of breast cancer. J. Exp. Clin. Cancer Res. 2023, 42, 343. [Google Scholar] [CrossRef]
- Ekanger, C.T.; Ramnefjell, M.P.; Guttormsen, M.S.; Hekland, J.; Dahl-Michelsen, K.; Lotsberg, M.L.; Lu, N.; Stuhr, L.E.; Hoareau, L.; Salminen, P.R.; et al. An Organoid Model for Translational Cancer Research Recapitulates Histoarchitecture and Molecular Hallmarks of Non-Small-Cell Lung Cancer. Cancers 2025, 17, 1873. [Google Scholar] [CrossRef] [PubMed]
- Nam, C.; Ziman, B.; Sheth, M.; Zhao, H.; Lin, D.C. Genomic and epigenomic characterization of tumor organoid models. Cancers 2022, 14, 4090. [Google Scholar] [CrossRef] [PubMed]
- El Harane, S.; Zidi, B.; El Harane, N.; Krause, K.H.; Matthes, T.; Preynat-Seauve, O. Cancer spheroids and organoids as novel tools for research and therapy: State of the art and challenges to guide precision medicine. Cells 2023, 12, 1001. [Google Scholar] [CrossRef] [PubMed]
- Monberg, M. Leveraging Single Cell Technologies for the Characterization and Treatment of Refractory Pancreatic Cancer. Ph.D. Thesis, The University of Texas, Houston, TX, USA, 2022. [Google Scholar]
- Xiao, Y.; Li, Y.; Jing, X.; Weng, L.; Liu, X.; Liu, Q.; Chen, K. Organoid models in oncology: Advancing precision cancer therapy and vaccine development. Cancer Biol. Med. 2025, 22, 903–927. [Google Scholar] [CrossRef]
- Marchini, A.; Gelain, F. Synthetic scaffolds for 3D cell cultures and organoids: Applications in regenerative medicine. Crit. Rev. Biotechnol. 2022, 42, 468–486. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef]
- Tu, S.M.; Singh, S.R.; Arnaoutakis, K.; Malapati, S.; Bhatti, S.A.; Joon, A.Y.; Atiq, O.T.; Pisters, L.L. Stem cell theory of cancer: Implications for translational research from bedside to bench. Cancers 2022, 14, 3345. [Google Scholar] [CrossRef]
- Pamarthy, S.; Sabaawy, H.E. Patient derived organoids in prostate cancer: Improving therapeutic efficacy in precision medicine. Mol. Cancer 2021, 20, 125. [Google Scholar] [CrossRef]
- Bisht, S.; Nigam, M.; Kunjwal, S.S.; Sergey, P.; Mishra, A.P.; Sharifi-Rad, J. Cancer stem cells: From an insight into the basics to recent advances and therapeutic targeting. Stem Cells Int. 2022, 2022, 9653244. [Google Scholar] [CrossRef]
- Ren, J.; Liu, M.; Rong, M.; Zhang, X.; Wang, G.; Liu, Y.; Li, H.; Duan, S. The pros and cons of mechanical dissociation and enzymatic digestion in patient-derived organoid cultures for solid tumor. Cell Organoid 2024, 1, 9410009. [Google Scholar] [CrossRef]
- Pang, X.; Hu, Y.; Dai, Z.; Lou, Q.; Xu, W.; Chen, L. Precision medicine research progress based on colorectal cancer organoids. Discov. Oncol. 2025, 16, 1181. [Google Scholar] [CrossRef] [PubMed]
- Alnasser, S.M. Advances and Challenges in Cancer Stem Cells for Onco-Therapeutics. Stem Cells Int. 2023, 2023, 8722803. [Google Scholar] [CrossRef] [PubMed]
- Izycka, N.; Rucinski, M.; Andrzejewska, M.; Szubert, S.; Nowak-Markwitz, E.; Sterzynska, K. The prognostic value of Cancer Stem Cell Markers (CSCs) expression—ALDH1A1, CD133, CD44—For survival and long-term follow-up of ovarian cancer patients. Int. J. Mol. Sci. 2023, 24, 2400. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; D’Souza, G.G. Modeling tumor microenvironment complexity in vitro: Spheroids as physiologically relevant tumor models and strategies for their analysis. Cells 2025, 14, 732. [Google Scholar] [CrossRef]
- Aldoghachi, A.F.; Chong, Z.X.; Yeap, S.K.; Cheong, S.K.; Ho, W.Y.; Ong, A.H. Stem cells for cancer therapy: Translating the uncertainties and possibilities of stem cell properties into opportunities for effective cancer therapy. Int. J. Mol. Sci. 2023, 24, 1012. [Google Scholar] [CrossRef]
- Cerneckis, J.; Cai, H.; Shi, Y. Induced pluripotent stem cells (iPSCs): Molecular mechanisms of induction and applications. Signal Transduct. Target. Ther. 2024, 9, 112. [Google Scholar] [CrossRef]
- Lo, Y.H.; Kolahi, K.S.; Du, Y.; Chang, C.Y.; Krokhotin, A.; Nair, A.; Sobba, W.D.; Karlsson, K.; Jones, S.J.; Longacre, T.A.; et al. A CRISPR/Cas9-engineered ARID1A-deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov. 2021, 11, 1562–1581. [Google Scholar] [CrossRef]
- Al-Kabani, A.; Huda, B.; Haddad, J.; Yousuf, M.; Bhurka, F.; Ajaz, F.; Patnaik, R.; Jannati, S.; Banerjee, Y. Exploring Experimental Models of Colorectal Cancer: A Critical Appraisal from 2D Cell Systems to Organoids, Humanized Mouse Avatars, Organ-on-Chip, CRISPR Engineering, and AI-Driven Platforms—Challenges and Opportunities for Translational Precision Oncology. Cancers 2025, 17, 2163. [Google Scholar]
- Miloradovic, D.; Pavlovic, D.; Jankovic, M.G.; Nikolic, S.; Papic, M.; Milivojevic, N.; Stojkovic, M.; Ljujic, B. Human embryos, induced pluripotent stem cells, and organoids: Models to assess the effects of environmental plastic pollution. Front. Cell Dev. Biol. 2021, 9, 709183. [Google Scholar] [CrossRef]
- Dave, B.; Tailor, J. Human stem cell models to unravel brain cancer. BMC Cancer 2024, 24, 1465. [Google Scholar] [CrossRef]
- Yehya, A.; Youssef, J.; Hachem, S.; Ismael, J.; Abou-Kheir, W. Tissue-specific cancer stem/progenitor cells: Therapeutic implications. World J. Stem Cells 2023, 15, 323. [Google Scholar] [CrossRef]
- Milne, J.V.; Mustafa, E.H.; Clemons, N.J. Modelling esophageal adenocarcinoma and Barrett’s esophagus with patient-derived organoids. Front. Mol. Biosci. 2024, 11, 1382070. [Google Scholar] [CrossRef]
- Zhou, J.; Shi, Y. Mesenchymal stem/stromal cells (MSCs): Origin, immune regulation, and clinical applications. Cell. Mol. Immunol. 2023, 20, 555–557. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Wu, Q.; Shang, B.; Zhang, K.; Zhang, W. Organoid co-culture models of the tumor microenvironment promote precision medicine. Cancer Innov. 2024, 3, e101. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.A. The role of circulating tumor cells as a liquid biopsy for cancer: Advances, biology, technical challenges, and clinical relevance. Cancers 2024, 16, 1377. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Wang, X.; Yang, C.; Fu, K.; Wang, F.; Fu, L. The culture and application of circulating tumor cell-derived organoids. Trends Cell Biol. 2025, 35, 364–380. [Google Scholar] [CrossRef]
- Marei, H.E. Stem Cell and Synthetic Embryo Models: Advances, Applications, and Ethical Considerations. Stem Cell Rev. Rep. 2025, 21, 1648–1668. [Google Scholar] [CrossRef]
- Azeez, S.S.; Hamad, R.S.; Hamad, B.K.; Shekha, M.S.; Bergsten, P. Advances in CRISPR-Cas technology and its applications: Revolutionising precision medicine. Front. Genome Ed. 2024, 6, 1509924. [Google Scholar] [CrossRef]
- Yan, H.H.N.; Chan, A.S.; Lai, F.P.; Leung, S.Y. Organoid cultures for cancer modeling. Cell Stem Cell 2023, 30, 917–937. [Google Scholar] [CrossRef] [PubMed]
- Verstegen, M.M.A.; Coppes, R.P.; Beghin, A.; De Coppi, P.; Gerli, M.F.M.; de Graeff, N.; Pan, Q.; Saito, Y.; Shi, S.; Zadpoor, A.A.; et al. Clinical applications of human organoids. Nat. Med. 2025, 31, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, X.; Zhang, J.; Zhang, J.; Yang, C.; Zhao, Y. Harnessing Organoid Platforms for Nanoparticle Drug Development. Drug Des. Dev. Ther. 2025, 19, 6125–6143. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- He, R.; Weng, Z.; Liu, Y.; Li, B.; Wang, W.; Meng, W.; Li, B.; Li, L. Application of Induced Pluripotent Stem Cells in Malignant Solid Tumors. Stem Cell Rev. Rep. 2023, 19, 2557–2575. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- De Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tao, X.; Zhu, J.; Dai, Z.; Du, Y.; Xie, Y.; Chu, X.; Fu, G.; Lei, Z. Tumor organoid-immune co-culture models: Exploring a new perspective of tumor immunity. Cell Death Discov. 2025, 11, 195. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Policastro, B.; Nissen, N.; Alves, C.L. Deciphering Breast Tumor Heterogeneity Through Patient-Derived Organoids and Circulating Tumor Cells. J. Pers. Med. 2025, 15, 271. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mukhare, R.; Gandhi, K.A.; Kadam, A.; Raja, A.; Singh, A.; Madhav, M.; Chaubal, R.; Pandey, S.; Gupta, S. Integration of Organoids With CRISPR Screens: A Narrative Review. Biol. Cell 2025, 117, e70006. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Badekila, A.K.; Kini, S.; Jaiswal, A.K. Fabrication techniques of biomimetic scaffolds in three-dimensional cell culture: A review. J. Cell. Physiol. 2021, 236, 741–762. [Google Scholar] [CrossRef]
- Yi, S.A.; Zhang, Y.; Rathnam, C.; Pongkulapa, T.; Lee, K.B. Bioengineering approaches for the advanced organoid research. Adv. Mater. 2021, 33, 2007949. [Google Scholar] [CrossRef]
- Valdoz, J.C.; Johnson, B.C.; Jacobs, D.J.; Franks, N.A.; Dodson, E.L.; Sanders, C.; Cribbs, C.G.; Van Ry, P.M. The ECM: To scaffold, or not to scaffold, that is the question. Int. J. Mol. Sci. 2021, 22, 12690. [Google Scholar] [CrossRef]
- Kozlowski, M.T.; Crook, C.J.; Ku, H.T. Towards organoid culture without Matrigel. Commun. Biol. 2021, 4, 1387. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, X.; Dowbaj, A.M.; Sljukic, A.; Bratlie, K.; Lin, L.; Fong, E.L.S.; Balachander, G.M.; Chen, Z.; Soragni, A.; et al. Organoids. Nat. Rev. Methods Prim. 2022, 2, 94. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rana, M.M.; De la Hoz Siegler, H. Evolution of hybrid hydrogels: Next-generation biomaterials for drug delivery and tissue engineering. Gels 2024, 10, 216. [Google Scholar] [CrossRef]
- Sharma, P.; Pal, V.K.; Roy, S. An overview of latest advances in exploring bioactive peptide hydrogels for neural tissue engineering. Biomater. Sci. 2021, 9, 3911–3938. [Google Scholar] [CrossRef]
- Thakuri, P.S.; Liu, C.; Luker, G.D.; Tavana, H. Biomaterials-Based Approaches to Tumor Spheroid and Organoid Modeling. Adv. Healthc. Mater. 2018, 7, e1700980. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shou, Y.; Teo, X.Y.; Wu, K.Z.; Bai, B.; Kumar, A.R.; Low, J.; Le, Z.; Tay, A. Dynamic stimulations with bioengineered extracellular matrix-mimicking hydrogels for mechano cell reprogramming and therapy. Adv. Sci. 2023, 10, 2300670. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, T.; Cui, H.; Cui, H. Integrating 3D bioprinting and organoids to better recapitulate the complexity of cellular microenvironments for tissue engineering. Adv. Healthc. Mater. 2025, 14, 2403762. [Google Scholar] [CrossRef] [PubMed]
- Smandri, A.; Al-Masawa, M.E.; Hwei, N.M.; Fauzi, M.B. ECM-derived biomaterials for regulating tissue multicellularity and maturation. iScience 2024, 27, 109141. [Google Scholar] [CrossRef]
- Scalise, M.; Marino, F.; Salerno, L.; Cianflone, E.; Molinaro, C.; Salerno, N.; De Angelis, A.; Viglietto, G.; Urbanek, K.; Torella, D. From spheroids to organoids: The next generation of model systems of human cardiac regeneration in a dish. Int. J. Mol. Sci. 2021, 22, 13180. [Google Scholar] [CrossRef]
- Liu, D.; Chen, S.; Win Naing, M. A review of manufacturing capabilities of cell spheroid generation technologies and future development. Biotechnol. Bioeng. 2021, 118, 542–554. [Google Scholar] [CrossRef]
- Arora, S.; Singh, S.; Mittal, A.; Desai, N.; Khatri, D.K.; Gugulothu, D.; Lather, V.; Pandita, D.; Vora, L.K. Spheroids in cancer research: Recent advances and opportunities. J. Drug Deliv. Sci. Technol. 2024, 100, 106033. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Wan, K.; Wu, M.; Guo, L.; Liu, X.; Wei, G. On the design, functions, and biomedical applications of high-throughput dielectrophoretic micro-/nanoplatforms: A review. Nanoscale 2021, 13, 4330–4358. [Google Scholar] [CrossRef]
- Garcia Garcia, F.C. Microfluidic Control of Oxygen for Long-Term Maintenance of Tissues on Chip. Ph.D. Thesis, University of Southampton, Southampton, UK, 2024. [Google Scholar]
- Picollet-D’hahan, N.; Zuchowska, A.; Lemeunier, I.; Le Gac, S. Multiorgan-on-a-chip: A systemic approach to model and decipher inter-organ communication. Trends Biotechnol. 2021, 39, 788–810. [Google Scholar] [CrossRef]
- Maramraju, S.; Kowalczewski, A.; Kaza, A.; Liu, X.; Singaraju, J.P.; Albert, M.V.; Ma, Z.; Yang, H. AI-organoid integrated systems for biomedical studies and applications. Bioeng. Transl. Med. 2024, 9, e10641. [Google Scholar] [CrossRef] [PubMed]
- Silva-Pedrosa, R.; Salgado, A.J.; Ferreira, P.E. Revolutionizing disease modeling: The emergence of organoids in cellular systems. Cells 2023, 12, 930. [Google Scholar] [CrossRef] [PubMed]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef] [PubMed]
- Giron-Michel, J.; Padelli, M.; Oberlin, E.; Guenou, H.; Duclos-Vallée, J.C. State-of-the-art liver cancer organoids: Modeling cancer stem cell heterogeneity for personalized treatment. BioDrugs 2025, 39, 237–260. [Google Scholar] [CrossRef]
- Huang, W.; Xu, Z.; Li, S.; Zhou, J.; Zhao, B. Living biobanks of organoids: Valuable resource for translational research. Biopreserv. Biobank. 2024, 22, 543–549. [Google Scholar] [CrossRef]
- Feng, Q.S.; Shan, X.F.; Yau, V.; Cai, Z.G.; Xie, S. Facilitation of Tumor Stroma-Targeted Therapy: Model Difficulty and Co-Culture Organoid Method. Pharmaceuticals 2025, 18, 62. [Google Scholar] [CrossRef]
- Gabriel, V.; Zdyrski, C.; Sahoo, D.K.; Ralston, A.; Wickham, H.; Bourgois-Mochel, A.; Ahmed, B.; Merodio, M.M.; Paukner, K.; Piñeyro, P.; et al. Adult animal stem cell-derived organoids in biomedical research and the one health paradigm. Int. J. Mol. Sci. 2024, 25, 701. [Google Scholar] [CrossRef]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Tufail, M.; Wu, C. Wnt3a is a promising target in colorectal cancer. Med. Oncol. 2023, 40, 86. [Google Scholar] [CrossRef] [PubMed]
- Indira Chandran, V.; Gopala, S.; Venkat, E.H.; Kjolby, M.; Nejsum, P. Extracellular vesicles in glioblastoma: A challenge and an opportunity. npj Precis. Oncol. 2024, 8, 103. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ornelas-González, A.; González-González, M.; Rito-Palomares, M. Microcarrier-based stem cell bioprocessing: GMP-grade culture challenges and future trends for regenerative medicine. Crit. Rev. Biotechnol. 2021, 41, 1081–1095. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Z.; Zhang, Y.; Zhong, H.; Cai, X.; Guan, R. Recent progress on the organoids: Techniques, advantages and applications. Biomed. Pharmacother. 2025, 185, 117942. [Google Scholar] [CrossRef]
- Lv, J.; Du, X.; Wang, M.; Su, J.; Wei, Y.; Xu, C. Construction of tumor organoids and their application to cancer research and therapy. Theranostics 2024, 14, 1101–1125. [Google Scholar] [CrossRef]
- Guzmán Herrera, A. Mechanics of Organoid Formation and Repair. Ph.D. Thesis, University College London, London, UK, 2023. [Google Scholar]
- Ulhaq, A. Organoid Computer Vision: A Survey and Outlook. engrXiv 2024. preprint. [Google Scholar]
- Man, Y.; Liu, Y.; Chen, Q.; Zhang, Z.; Li, M.; Xu, L.; Tan, Y.; Liu, Z. Organoids-On-a-Chip for Personalized Precision Medicine. Adv. Healthc. Mater. 2024, 13, 2401843. [Google Scholar] [CrossRef]
- Uhrig, M.; Ezquer, F.; Ezquer, M. Improving cell recovery: Freezing and thawing optimization of induced pluripotent stem cells. Cells 2022, 11, 799. [Google Scholar] [CrossRef]
- Ahn, S.J. Standards for organoids. Int. J. Stem Cells 2024, 17, 99–101. [Google Scholar] [CrossRef]
- Veninga, V.; Voest, E.E. Tumor organoids: Opportunities and challenges to guide precision medicine. Cancer Cell 2021, 39, 1190–1201. [Google Scholar] [CrossRef]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.W.; Min, D.W.; Kim, H.P.; An, Y.; Kim, S.; Youk, J.; Chun, J.; Im, J.P.; Song, S.H.; Ju, Y.S.; et al. Patient-derived organoids as a preclinical platform for precision medicine in colorectal cancer. Mol. Oncol. 2022, 16, 2396–2412. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ren, H.; Jia, X.; Yu, L. The building blocks of embryo models: Embryonic and extraembryonic stem cells. Cell Discov. 2025, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Romanesco, R. The Added Value of Organoid Models in the Context of Pediatric Cancer Research. Master’s Thesis, Utrecht University, Utrecht, The Netherlands, 2025. [Google Scholar]
- Hsiung, N.; Ju, Y.; Yang, K.; Yang, P.; Zeng, W.; Zhao, H.; Zou, P.; Ye, J.; Yi, K.; Wang, X. Organoid-based tissue engineering for advanced tissue repair and reconstruction. Mater. Today Bio 2025, 33, 102093. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Seidlitz, T.; Stange, D.E. Gastrointestinal cancer organoids-applications in basic and translational cancer research. Exp. Mol. Med. 2021, 53, 1459–1470. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, W.; Zhou, Z.; Zhou, X.; Khoo, B.L.; Gunawan, R.; Chin, Y.R.; Zhang, L.; Yi, C.; Guan, X.; Yang, M. 3D biomimetic models to reconstitute tumor microenvironment in vitro: Spheroids, organoids, and tumor-on-a-chip. Adv. Healthc. Mater. 2023, 12, 2202609. [Google Scholar] [CrossRef]
- Praharaj, P.P.; Bhutia, S.K.; Nagrath, S.; Bitting, R.L.; Deep, G. Circulating tumor cell-derived organoids: Current challenges and promises in medical research and precision medicine. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 117–127. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bhattacharya, R.; Bose, D.; Kaur, T.; Patel, R.; Renuka, O.; Rodriguez, R.V. Model organoids: Integrated frameworks for the next frontier of healthcare advancements. Stem Cell Rev. Rep. 2025, 21, 319–336. [Google Scholar] [CrossRef]
- Ballav, S.; Deshmukh, A.J.; Siddiqui, S.; Aich, J.; Basu, S. Two-dimensional and three-dimensional cell culture and their applications. In Cell Culture-Advanced Technology and Applications in Medical and Life Sciences; IntechOpen: Rijeka, Croatia, 2021. [Google Scholar]
- Ćosić, M.; Petelinec, A. The role of 3D cell cultures in understanding mitosis and tissue architecture. Period. Biol. 2024, 126, 1–4. [Google Scholar] [CrossRef]
- Abbas, Z.N.; Al-Saffar, A.Z.; Jasim, S.M.; Sulaiman, G.M. Comparative analysis between 2D and 3D colorectal cancer culture models for insights into cellular morphological and transcriptomic variations. Sci. Rep. 2023, 13, 18380. [Google Scholar] [CrossRef]
- Valkenburg, K.C.; de Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Colombo, E.; Cattaneo, M.G. Multicellular 3D models to study tumour-stroma interactions. Int. J. Mol. Sci. 2021, 22, 1633. [Google Scholar] [CrossRef]
- Gayan, S.; Teli, A.; Nair, A.; Tomar, G.; Dey, T. Macro-and micro-nutrient–based multiplex stress conditions modulate in vitro tumorigenesis and aggressive behavior of breast cancer spheroids. Vitr. Model. 2022, 1, 85–101. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D in vitro model (R) evolution: Unveiling tumor–stroma interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.; Lee, H.J. Spheroid-Hydrogel-Integrated Biomimetic System: A New Frontier in Advanced Three-Dimensional Cell Culture Technology. Cells Tissues Organs 2025, 214, 128–147. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jin, K.; Teng, L.; Shen, Y.; He, K.; Xu, Z.; Li, G. Patient-derived human tumour tissue xenografts in immunodeficient mice: A systematic review. Clin. Transl. Oncol. 2010, 12, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, Z.; Brown, E.A.; Ghazaryan, A.; Welm, A.L. PDX models for functional precision oncology and discovery science. Nat. Rev. Cancer 2025, 25, 153–166. [Google Scholar] [CrossRef]
- Chiorazzi, M.; Martinek, J.; Krasnick, B.; Zheng, Y.; Robbins, K.J.; Qu, R.; Kaufmann, G.; Skidmore, Z.; Juric, M.; Henze, L.A.; et al. Autologous humanized PDX modeling for immuno-oncology recapitulates features of the human tumor microenvironment. J. Immunother. Cancer 2023, 11, e006921. [Google Scholar] [CrossRef]
- Cammarota, F.; Laukkanen, M.O. Mesenchymal stem/stromal cells in stromal evolution and cancer progression. Stem Cells Int. 2016, 2016, 4824573. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, F.; Jin, Y.; Ma, Y. Applications of human organoids in the personalized treatment for digestive diseases. Signal Transduct. Target. Ther. 2022, 7, 336. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, X.; Francies, H.E.; Secrier, M.; Perner, J.; Miremadi, A.; Galeano-Dalmau, N.; Barendt, W.J.; Letchford, L.; Leyden, G.M.; Goffin, E.K.; et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018, 9, 2983. [Google Scholar] [CrossRef] [PubMed]
- Gunti, S.; Hoke, A.T.; Vu, K.P.; London, N.R., Jr. Organoid and spheroid tumor models: Techniques and applications. Cancers 2021, 13, 874. [Google Scholar] [CrossRef] [PubMed]
- Ronzetti, M.; Simeonov, A. A comprehensive update on the application of high-throughput fluorescence imaging for novel drug discovery. Expert Opin. Drug Discov. 2025, 20, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Fedi, A.; Vitale, C.; Giannoni, P.; Caluori, G.; Marrella, A. Biosensors to monitor cell activity in 3D hydrogel-based tissue models. Sensors 2022, 22, 1517. [Google Scholar] [CrossRef]
- de la Rocha, A.M.; Berlow, N.E.; Azzam, D.J. Functional precision medicine: The future of cancer care. Trends Mol. Med. 2025, 31, 404–408. [Google Scholar] [CrossRef]
- Aksoy, S.A.; Earl, J.; Grahovac, J.; Karakas, D.; Lencioni, G.; Sığırlı, S.; Bijlsma, M.F. Organoids, tissue slices and organotypic cultures: Advancing our understanding of pancreatic ductal adenocarcinoma through in vitro and ex vivo models. Semin. Cancer Biol. 2025, 109, 10–24. [Google Scholar] [CrossRef]
- Liu, F.; Li, G.; Zheng, Y.; Liu, Y.; Liu, K. Multiplex imaging analysis of the tumor immune microenvironment for guiding precision immunotherapy. Front. Immunol. 2025, 16, 1617906. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Lin, H.; Zhu, Y.; Huang, D.; Lai, M.; Xi, X.; Huang, J.; Zhang, W.; Zhong, T. Research progress of CTC, ctDNA, and EVs in cancer liquid biopsy. Front. Oncol. 2024, 14, 1303335. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Beyes, S.; Bediaga, N.G.; Zippo, A. An epigenetic perspective on intra-tumour heterogeneity: Novel insights and new challenges from multiple fields. Cancers 2021, 13, 4969. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Pelosi, E.; Castelli, G. Colorectal cancer: Genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med. Sci. 2018, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. Phenotypic heterogeneity, bidirectionality, universal cues, plasticity, mechanics, and the tumor microenvironment drive cancer metastasis. Biomolecules 2024, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jiang, M.; Wang, H.; Sun, H.; Zhu, J.; Zhao, W.; Fang, Q.; Yu, J.; Chen, P.; Wu, S.; et al. A narrative review of tumor heterogeneity and challenges to tumor drug therapy. Ann. Transl. Med. 2021, 9, 1351. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, P.; Wang, J.; Lv, H.; Han, J.; Hou, Z.; Xu, R.; Chen, W. Advances in spatial transcriptomics and its application in the musculoskeletal system. Bone Res. 2025, 13, 54. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fu, Y.C.; Liang, S.B.; Luo, M.; Wang, X.P. Intratumoral heterogeneity and drug resistance in cancer. Cancer Cell Int. 2025, 25, 103. [Google Scholar] [CrossRef]
- Seppälä, T.T.; Zimmerman, J.W.; Sereni, E.; Plenker, D.; Suri, R.; Rozich, N.; Blair, A.; Thomas, D.L.; Teinor, J.; Javed, A.; et al. Patient-derived organoid pharmacotyping is a clinically tractable strategy for precision medicine in pancreatic cancer. Ann. Surg. 2020, 272, 427–435. [Google Scholar] [CrossRef]
- Wagner, D.E.; Klein, A.M. Lineage tracing meets single-cell omics: Opportunities and challenges. Nat. Rev. Genet. 2020, 21, 410–427. [Google Scholar] [CrossRef]
- Yang, Y.; Li, S.; Wang, Y.; Zhao, Y.; Li, Q. Protein tyrosine kinase inhibitor resistance in malignant tumors: Molecular mechanisms and future perspective. Signal Transduct. Target. Ther. 2022, 7, 329. [Google Scholar] [CrossRef]
- Wang, R.C.; Wang, Z. Precision medicine: Disease subtyping and tailored treatment. Cancers 2023, 15, 3837. [Google Scholar] [CrossRef]
- James, C.; Whitehead, A.; Plummer, J.T.; Thompson, R.; Badal, S. Failure to progress: Breast and prostate cancer cell lines in developing targeted therapies. Cancer Metastasis Rev. 2024, 43, 1529–1548. [Google Scholar] [CrossRef] [PubMed]
- Sar, F.; Chung, H.C.; Lin, Y.Y.; Lin, D.; Morova, T.; Haegert, A.; Volik, S.; Bell, R.; LeBihan, S.; Ozturan, D.; et al. Longitudinal single-cell RNA sequencing of a neuroendocrine transdifferentiation model reveals transcriptional reprogramming in treatment-induced neuroendocrine prostate cancer. bioRxiv 2025. preprint. [Google Scholar]
- Zhou, Z.; Cong, L.; Cong, X. Patient-derived organoids in precision medicine: Drug screening, organoid-on-a-chip and living organoid biobank. Front. Oncol. 2021, 11, 762184. [Google Scholar] [CrossRef] [PubMed]
- Tonsing-Carter, E.; Agarwal, R.; Kyi, C.W.; Perez-Mayoral, J.; Soria, C.T.; Zenklusen, J.C. Human cancer models initiative (HCMI): A community resource of next-generation cancer models and associated data. Cancer Res. 2023, 83 (Suppl. S7), 4681. [Google Scholar] [CrossRef]
- Sanger Institute. Organoid Cell Lines—Cancer Models; Wellcome Trust Sanger Institute: Cambridge, UK, 2023; Available online: https://www.sanger.ac.uk/technology/organoid-cell-lines/ (accessed on 4 February 2019).
- Li, H.; Liu, H.; Chen, K. Living biobank-based cancer organoids: Prospects and challenges in cancer research. Cancer Biol. Med. 2022, 19, 965–982. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, B.; Luo, F.; Guo, Y.; Jiang, N.; Wei, J.; Wang, X.; Tseng, Y.; Chen, J.; Zhao, B.; et al. Establishment of a large-scale patient-derived high-risk colorectal adenoma organoid biobank for high-throughput and high-content drug screening. BMC Med. 2023, 21, 336. [Google Scholar] [CrossRef]
- Brancato, V.; Esposito, G.; Coppola, L.; Cavaliere, C.; Mirabelli, P.; Scapicchio, C.; Borgheresi, R.; Neri, E.; Salvatore, M.; Aiello, M. Standardizing digital biobanks: Integrating imaging, genomic, and clinical data for precision medicine. J. Transl. Med. 2024, 22, 136. [Google Scholar] [CrossRef]
- Mukherjee, A.; Abraham, S.; Singh, A.; Balaji, S.; Mukunthan, K.S. From data to cure: A comprehensive exploration of multi-omics data analysis for targeted therapies. Mol. Biotechnol. 2025, 67, 1269–1289. [Google Scholar] [CrossRef]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef]
- Grüner, B.M.; Schulze, C.J.; Yang, D.; Ogasawara, D.; Dix, M.M.; Rogers, Z.N.; Chuang, C.H.; McFarland, C.D.; Chiou, S.H.; Brown, J.M.; et al. An in vivo multiplexed small-molecule screening platform. Nat. Methods 2016, 13, 883–889. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tong, L.; Cui, W.; Zhang, B.; Fonseca, P.; Zhao, Q.; Zhang, P.; Xu, B.; Zhang, Q.; Li, Z.; Seashore-Ludlow, B.; et al. Patient-derived organoids in precision cancer medicine. Med 2024, 5, 1351–1377. [Google Scholar] [CrossRef]
- Dilmac, S.; Ozpolat, B. Mechanisms of PARP-Inhibitor-Resistance in BRCA-Mutated Breast Cancer and New Therapeutic Approaches. Cancers 2023, 15, 3642. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bironzo, P.; Reale, M.L.; Sperone, T.; Tabbò, F.; Caglio, A.; Listì, A.; Passiglia, F.; Di Maio, M.; Righi, L.; Bussolino, F.; et al. Clinical and molecular features of epidermal growth factor receptor (EGFR) mutation positive non-small-cell lung cancer (NSCLC) patients treated with tyrosine kinase inhibitors (TKIs): Predictive and prognostic role of co-mutations. Cancers 2021, 13, 2425. [Google Scholar] [CrossRef] [PubMed]
- Morel, D.; Jeffery, D.; Aspeslagh, S.; Almouzni, G.; Postel-Vinay, S. Combining epigenetic drugs with other therapies for solid tumours—Past lessons and future promise. Nat. Rev. Clin. Oncol. 2020, 17, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Tardito, S.; Matis, S.; Zocchi, M.R.; Benelli, R.; Poggi, A. Epidermal growth factor receptor targeting in colorectal carcinoma: Antibodies and patient-derived organoids as a smart model to study therapy resistance. Int. J. Mol. Sci. 2024, 25, 7131. [Google Scholar] [CrossRef] [PubMed]
- Cicolini, M. Development of a Microfluidic Alveolus-on-Chip Supporting Epithelial-Endothelial Cells Co-Culture and Air-Liquid Interface Implementation for the Modelling of the Physiological Alveolar-Capillary Barrier. Ph.D. Thesis, Politecnico di Torino, Torino, Italy, 2022. [Google Scholar]
- Parseh, B.; Khosravi, A.; Fazel, A.; Ai, J.; Ebrahimi-Barough, S.; Verdi, J.; Shahbazi, M. 3-Dimensional model to study apoptosis induction of activated natural killer cells conditioned medium using patient-derived colorectal cancer organoids. Front. Cell Dev. Biol. 2022, 10, 895284. [Google Scholar] [CrossRef]
- Li, P.; Huang, M.; Li, M.; Li, G.; Ma, Y.; Zhao, Y.; Wang, X.; Zhang, Y.; Shi, C. Combining molecular characteristics and therapeutic analysis of PDOs predict clinical responses and guide PDAC personalized treatment. J. Exp. Clin. Cancer Res. 2025, 44, 72. [Google Scholar] [CrossRef]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef]
- Guedan, S.; Luu, M.; Ammar, D.; Barbao, P.; Bonini, C.; Bousso, P.; Buchholz, C.J.; Casucci, M.; De Angelis, B.; Donnadieu, E.; et al. Time 2EVOLVE: Predicting efficacy of engineered T-cells–how far is the bench from the bedside? J. Immunother. Cancer 2022, 10, e003487. [Google Scholar] [CrossRef]
- D’Accardo, C. The Use of CD44v6-CAR T Cells in Combination with PD-1/PD-L1 and PI3K/AKT Pathway Inhibitors as Therapeutic Strategies for Advanced Cancer. Ph.D. Thesis, University of Palermo, Palermo, Italy, 2024. [Google Scholar]
- Palacios, P.A.; Flores, I.; Cereceda, L.; Otero, F.F.; Müller, M.; Brebi, P.; Contreras, H.R.; Carreño, L.J. Patient-Derived Organoid Models for NKT Cell-Based Cancer Immunotherapy. Cancers 2025, 17, 406. [Google Scholar] [CrossRef]
- Lee, R.Y.; Wu, Y.; Goh, D.; Tan, V.; Ng, C.W.; Lim, J.C.; Lau, M.C.; Yeong, J.P. Application of artificial intelligence to in vitro tumor modeling and characterization of the tumor microenvironment. Adv. Healthc. Mater. 2023, 12, 2202457. [Google Scholar] [CrossRef]
- Polak, R.; Zhang, E.T.; Kuo, C.J. Cancer organoids 2.0: Modelling the complexity of the tumour immune microenvironment. Nat. Rev. Cancer 2024, 24, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Murias-Closas, A.; Prats, C.; Calvo, G.; López-Codina, D.; Olesti, E. Computational modelling of CAR T-cell therapy: From cellular kinetics to patient-level predictions. eBioMedicine 2025, 113, 105597. [Google Scholar] [CrossRef] [PubMed]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef] [PubMed]
- Bruna, A.; Rueda, O.M.; Greenwood, W.; Batra, A.S.; Callari, M.; Batra, R.N.; Pogrebniak, K.; Sandoval, J.; Cassidy, J.W.; Tufegdzic-Vidakovic, A.; et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell 2016, 167, 260–274.e22. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, Y.; Chen, P. Lung cancer organoids: Models for preclinical research and precision medicine. Front. Oncol. 2023, 13, 1293441. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pan, Y.; Cui, H.; Song, Y. Organoid drug screening report for a non-small cell lung cancer patient with EGFR gene mutation negativity: A case report and review of the literature. Front. Oncol. 2023, 13, 1109274. [Google Scholar] [CrossRef]
- Napoli, G.C.; Figg, W.D.; Chau, C.H. Functional drug screening in the era of precision medicine. Front. Med. 2022, 9, 912641. [Google Scholar] [CrossRef]
- Miserocchi, G.; Bocchini, M.; Cortesi, M.; Arienti, C.; De Vita, A.; Liverani, C.; Mercatali, L.; Bravaccini, S.; Ulivi, P.; Zanoni, M. Combining preclinical tools and models to unravel tumor complexity: Jump into the next dimension. Front. Immunol. 2023, 14, 1171141. [Google Scholar] [CrossRef]
- Jeong, Y.K.; Song, B.; Bae, S. Current status and challenges of DNA base editing tools. Mol. Ther. 2020, 28, 1938–1952. [Google Scholar] [CrossRef]
- Ferreira, A.; Pereira, F.; Reis, C.; Oliveira, M.J.; Sousa, M.J.; Preto, A. Crucial role of oncogenic KRAS mutations in apoptosis and autophagy regulation: Therapeutic implications. Cells 2022, 11, 2183. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Ge, H.; Ghadban, T.; Reeh, M.; Güngör, C. The extracellular matrix: A key accomplice of cancer stem cell migration, metastasis formation, and drug resistance in PDAC. Cancers 2022, 14, 3998. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Guo, Z.; Xu, X.; Li, P.; Fang, Y.; Deng, S. Advances in CRISPR-Cas9 in lineage tracing of model animals. Anim. Model. Exp. Med. 2025, 8, 1004–1022. [Google Scholar] [CrossRef] [PubMed]
- Soragni, A.; Knudsen, E.S.; O’Connor, T.N.; Tognon, C.E.; Tyner, J.W.; Gini, B.; Kim, D.; Bivona, T.G.; Zang, X.; Witkiewicz, A.K.; et al. Acquired resistance in cancer: Towards targeted therapeutic strategies. Nat. Rev. Cancer 2025, 25, 613–633. [Google Scholar] [CrossRef] [PubMed]
- Al-Qadami, G.; Raposo, A.; Chien, C.C.; Ma, C.; Priebe, I.; Hor, M.; Fung, K. Intestinal organoid coculture systems: Current approaches, challenges, and future directions. Am. J. Physiol. Gastrointest. Liver Physiol. 2025, 328, G252–G276. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Zeevi, D.; Dohnalová, L.; Zilberman-Schapira, G.; Mahdi, J.A.; David, E.; Savidor, A.; Korem, T.; Herzig, Y.; et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 2015, 163, 1428–1443. [Google Scholar] [CrossRef]
- Monteiro, C.F.; Deus, I.A.; Custódio, C.A.; Mano, J.F. Biomaterials meet organ-on-chips–a perspective on tumor modeling. Int. Mater. Rev. 2025, 70, 31–68. [Google Scholar] [CrossRef]
- Schilderink, R.; Verseijden, C.; Seppen, J.; Muncan, V.; Van Den Brink, G.R.; Lambers, T.T.; van Tol, E.A.; de Jonge, W.J. The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G1138–G1146. [Google Scholar] [CrossRef]
- Iftekhar, A.; Berger, H.; Bouznad, N.; Heuberger, J.; Boccellato, F.; Dobrindt, U.; Hermeking, H.; Sigal, M.; Meyer, T.F. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat. Commun. 2021, 12, 1003. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Strating, E.; Verhagen, M.P.; Wensink, E.; Dünnebach, E.; Wijler, L.; Aranguren, I.; De la Cruz, A.S.; Peters, N.A.; Hageman, J.H.; van der Net, M.M.; et al. Co-cultures of colon cancer cells and cancer-associated fibroblasts recapitulate the aggressive features of mesenchymal-like colon cancer. Front. Immunol. 2023, 14, 1053920. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, H.; Luo, X.; Chen, Y.; Shi, C.; Wang, Y.; Bai, J.; Shao, Z.; Shang, Z. NNMT switches the proangiogenic phenotype of cancer-associated fibroblasts via epigenetically regulating ETS2/VEGFA axis. Oncogene 2024, 43, 2647–2660. [Google Scholar] [CrossRef] [PubMed]
- Rosales, A.M.; Anseth, K.S. The design of reversible hydrogels to capture extracellular matrix dynamics. Nat. Rev. Mater. 2016, 1, 15012. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.M.; Deka, D.; Das, A.; Paul, S.; Pathak, S.; Banerjee, A. Role of interleukins in inflammation-mediated tumor immune microenvironment modulation in colorectal cancer pathogenesis. Dig. Dis. Sci. 2023, 68, 3220–3236. [Google Scholar] [CrossRef] [PubMed]
- Atiya, H.; Frisbie, L.; Pressimone, C.; Coffman, L. Mesenchymal stem cells in the tumor microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 31–42. [Google Scholar]
- Fernando, K.; Kwang, L.G.; Lim, J.T.; Fong, E.L. Hydrogels to engineer tumor microenvironments in vitro. Biomater. Sci. 2021, 9, 2362–2383. [Google Scholar] [CrossRef]
- Jia, H.; Chen, X.; Zhang, L.; Chen, M. Cancer associated fibroblasts in cancer development and therapy. J. Hematol. Oncol. 2025, 18, 36. [Google Scholar] [CrossRef]
- Esparza, A.; Jimenez, N.; Borrego, E.A.; Browne, S.; Natividad-Diaz, S.L. Review: Human stem cell-based 3D in vitro angiogenesis models for preclinical drug screening applications. Mol. Biol. Rep. 2024, 51, 260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, R.; Brislinger, D.; Fuchs, J.; Lyons, A.; Langthaler, S.; Hauser, C.A.E.; Baumgartner, C. Vascularised organoids: Recent advances and applications in cancer research. Clin. Transl. Med. 2025, 15, e70258. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dong, E.; Yue, X.Z.; Shui, L.; Liu, B.R.; Li, Q.Q.; Yang, Y.; Luo, H.; Wang, W.; Yang, H.S. IFN-γ surmounts PD-L1/PD1 inhibition to CAR-T cell therapy by upregulating ICAM-1 on tumor cells. Signal Transduct. Target. Ther. 2021, 6, 20. [Google Scholar] [CrossRef]
- Djomehri, S.I.; Burman, B.; Gonzalez, M.E.; Takayama, S.; Kleer, C.G. A reproducible scaffold-free 3D organoid model to study neoplastic progression in breast cancer. J. Cell Commun. Signal. 2019, 13, 129–143. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, Z.; Yue, M.; Liu, Y.; Zhang, P.; Qing, J.; Liu, H.; Zhou, Y. Advances of Engineered Hydrogel Organoids within the Stem Cell Field: A Systematic Review. Gels 2022, 8, 379. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, K.; Chen, X.; Fan, Z.; Ren, F.; Liu, J.; Hu, B. From organoids to organoids-on-a-chip: Current applications and challenges in biomedical research. Chin. Med. J. 2025, 138, 792–807. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Subtil, B.; Iyer, K.K.; Poel, D.; Bakkerus, L.; Gorris, M.A.J.; Escalona, J.C.; van den Dries, K.; Cambi, A.; Verheul, H.M.W.; de Vries, I.J.M.; et al. Dendritic cell phenotype and function in a 3D co-culture model of patient-derived metastatic colorectal cancer organoids. Front. Immunol. 2023, 14, 1105244. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gaebler, D.; Hachey, S.J.; Hughes, C.C.W. Improving tumor microenvironment assessment in chip systems through next-generation technology integration. Front. Bioeng. Biotechnol. 2024, 12, 1462293. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Richiardone, E.; Van den Bossche, V.; Corbet, C. Metabolic studies in organoids: Current applications, opportunities and challenges. Organoids 2022, 1, 85–105. [Google Scholar] [CrossRef]
- Derouane, F.; Desgres, M.; Moroni, C.; Ambroise, J.; Berlière, M.; Van Bockstal, M.R.; Galant, C.; van Marcke, C.; Vara-Messler, M.; Hutten, S.J.; et al. Metabolic adaptation towards glycolysis supports resistance to neoadjuvant chemotherapy in early triple negative breast cancers. Breast Cancer Res. 2024, 26, 29. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Gevers, S.; Kok, R.N.; Burgering, L.M.; Neikes, H.; Akkerman, N.; Betjes, M.A.; Ludikhuize, M.C.; Gulersonmez, C.; Stigter, E.C.; et al. Lactate controls cancer stemness and plasticity through epigenetic regulation. Cell Metab. 2025, 37, 903–919.e10. [Google Scholar] [CrossRef]
- Andrews, M.G.; Kriegstein, A.R. Challenges of organoid research. Annu. Rev. Neurosci. 2022, 45, 23–39. [Google Scholar] [CrossRef]
- ElHarouni, D.; Al-Jazrawe, M.; Choi, S.; Dede, M.; Hinoue, T.; Misek, S.A.; Noh, H.; Zanella, L.; Tseng, M.; Francies, H.E.; et al. Abstract B025: Integrative clinical and molecular analysis of 665 next-generation in vitro cancer models generated by the the Human Cancer Models Initiative (HCMI) for advancing precision medicine and functional drug discovery. Cancer Res. 2025, 85 (Suppl. S5), B025. [Google Scholar] [CrossRef]
- Li, K.; He, Y.; Jin, X.; Jin, K.; Qian, J. Reproducible extracellular matrices for tumor organoid culture: Challenges and opportunities. J. Transl. Med. 2025, 23, 497. [Google Scholar] [CrossRef] [PubMed]
- Magno, V.; Meinhardt, A.; Werner, C. Polymer hydrogels to guide organotypic and organoid cultures. Adv. Funct. Mater. 2020, 30, 2000097. [Google Scholar] [CrossRef]
- Cheruku, G.R.; Wilson, C.V.; Raviendran, S.; Xiao, Q. Recent Advances and Future Perspectives in Vascular Organoids and Vessel-on-Chip. Organoids 2024, 3, 203–246. [Google Scholar] [CrossRef]
- Goenka, A.; Khan, F.; Verma, B.; Sinha, P.; Dmello, C.C.; Jogalekar, M.P.; Gangadaran, P.; Ahn, B.C. Tumor microenvironment signaling and therapeutics in cancer progression. Cancer Commun. 2023, 43, 525–561. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Morrison, A.I.; Sjoerds, M.J.; Vonk, L.A.; Gibbs, S.; Koning, J.J. In vitro immunity: An overview of immunocompetent organ-on-chip models. Front. Immunol. 2024, 15, 1373186. [Google Scholar] [CrossRef]
- Louey, A.; Hernández, D.; Pébay, A.; Daniszewski, M. Automation of organoid cultures: Current protocols and applications. SLAS Discov. Adv. Sci. Drug Discov. 2021, 26, 1138–1147. [Google Scholar] [CrossRef]
- Pridgeon, C.S. Development of Stem Cell-Derived Hepatocyte Models. Ph.D. Thesis, The University of Liverpool, Liverpool, UK, 2019. [Google Scholar]
- van der Velden, D.L.; Hoes, L.R.; van der Wijngaart, H.; van Berge Henegouwen, J.M.; van Werkhoven, E.; Roepman, P.; Schilsky, R.L.; de Leng, W.W.; Huitema, A.D.; Nuijen, B.; et al. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature 2019, 574, 127–131. [Google Scholar] [CrossRef]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor organoids as a pre-clinical cancer model for drug discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef]
- De Jongh, D.; Massey, E.K.; Bunnik, E.M. Organoids: A systematic review of ethical issues. Stem Cell Res. Ther. 2022, 13, 337. [Google Scholar] [CrossRef]
- Yu, F.; Hunziker, W.; Choudhury, D. Engineering microfluidic organoid-on-a-chip platforms. Micromachines 2019, 10, 165. [Google Scholar] [CrossRef]
- Baptista, L.S.; Porrini, C.; Kronemberger, G.S.; Kelly, D.J.; Perrault, C.M. 3D organ-on-a-chip: The convergence of microphysiological systems and organoids. Front. Cell Dev. Biol. 2022, 10, 1043117. [Google Scholar] [CrossRef]
- Clevenger, A.J.; McFarlin, M.K.; Collier, C.A.; Sheshadri, V.S.; Madyastha, A.K.; Gorley, J.P.; Solberg, S.C.; Stratman, A.N.; Raghavan, S.A. Peristalsis-associated mechanotransduction drives malignant progression of colorectal cancer. Cell. Mol. Bioeng. 2023, 16, 261–281. [Google Scholar] [CrossRef]
- Picco, G.; Rao, Y.; Al Saedi, A.; Lee, Y.; Vieira, S.F.; Bhosle, S.; May, K.; Herranz-Ors, C.; Walker, S.J.; Shenje, R.; et al. Novel WRN helicase inhibitors selectively target microsatellite-unstable cancer cells. Cancer Discov. 2024, 14, 1457–1475. [Google Scholar] [CrossRef]
- Psilopatis, I.; Sykaras, A.G.; Mandrakis, G.; Vrettou, K.; Theocharis, S. Patient-derived organoids: The beginning of a new era in ovarian cancer disease modeling and drug sensitivity testing. Biomedicines 2022, 11, 1. [Google Scholar] [CrossRef]

| Stem Cell Source | Biological Characteristics | Emerging Oncological Applications | Recent Innovations and Developments | Source |
|---|---|---|---|---|
| Adult Stem Cells (ASCs) | Tissue-resident, lineage-restricted; retain tumor-specific mutations and architecture. | Generation of patient-derived organoids (PDOs), drug screening, and therapy response prediction | Improved culture media for rare cancers; biobanking and automation for personalized oncology platforms | [30,51,52] |
| Cancer Stem Cells (CSCs) | Tumor-initiating, chemoresistant, and self-renewing; key drivers of heterogeneity and relapse | Modeling tumor recurrence, metastasis, and drug resistance evolution | Single-cell CSC organoid derivation; metabolic and immune-resistance profiling for precision immunotherapy | [34,53] |
| Induced Pluripotent Stem Cells (iPSCs) | Reprogrammed from somatic cells; pluripotent with epigenetic memory in some cases. | Controlled tumor initiation models, gene-function studies, and rare/pediatric cancer modeling | CRISPR-edited isogenic iPSC organoids are used in synthetic lethality screens and mutation-specific drug discovery. | [38,54] |
| Embryonic Stem Cells (ESCs) | Pluripotent, high differentiation potential; unrestricted developmental capacity | Modeling congenital tumors, developmental toxicology, pediatric brain and liver cancers | ESC-derived tumor organoids used in modeling syndromic mutations (e.g., DICER1); new ethical protocols in development | [41,42] |
| Tissue-Resident Progenitors | Committed progenitors from pre-malignant or inflamed tissues; transformation-prone | Modeling early-stage carcinogenesis (e.g., IBD-associated CRC, Barrett’s esophagus) | Used to study field cancerization, inflammation-driven transformation, and chromosomal instability (CIN) | [44,55] |
| Mesenchymal Stem Cells (MSCs) | Multipotent stromal cells with immunomodulatory and ECM remodeling capabilities | Tumor–stromal co-culture systems, angiogenesis, and immune evasion modeling | MSC–tumor organoid co-cultures for immune checkpoint therapy prediction; MSCs as vehicles for cytokine delivery in chips | [45,46,56] |
| Circulating Tumor Cells (CTCs) | Shed from primary tumors; highly metastatic; accessible via liquid biopsy | Real-time tumor modeling, tracking clonal evolution, and non-invasive resistance profiling | CTC-derived organoids used in breast, prostate, and lung cancer for longitudinal therapy monitoring | [47,48,57] |
| Engineered Synthetic Progenitors | Custom-designed using genetic circuits or modular engineering from iPSCs/ESCs | Functional genomics, mutation–phenotype correlation, scalable cancer modeling | Synthetic organoid libraries created for multiplexed CRISPR screening; ideal for identifying context-specific vulnerabilities | [49,50,58] |
| Organoid System Type | Primary Cellular Source | Scaffold Configuration | Media Composition and Microenvironmental Factors | Scalability and Throughput Potential | Cancer Models Addressed | Emerging Applications and Technological Innovations | Principal Limitations and Challenges | Source |
|---|---|---|---|---|---|---|---|---|
| ASC-Derived Organoids | Tumor-resident adult stem cells isolated from patient biopsies | Natural ECM hydrogels (e.g., Matrigel, collagen I) | Defined media enriched with EGF, Wnt3a, R-spondin, Noggin, A83-01 to sustain stemness | Moderate; supports batch production and cryopreservation | Colorectal, gastric, pancreatic, breast, prostate, lung | Large-scale patient-derived organoid (PDO) biobanking, AI-integrated drug sensitivity profiling, personalized therapy development | Variability in ECM batches; limited integration of stromal and immune components | [95,96] |
| CSC-Enriched Organoids | Purified cancer stem cells (e.g., CD44+, ALDH1+ subpopulations) | Suspension cultures or ECM-based matrices | Stemness-maintaining media with minimized differentiation cues to preserve CSC phenotypes | Low to moderate; challenging to expand in large quantities | Glioblastoma, triple-negative breast cancer, hepatocellular carcinoma, ovarian | Mapping of drug resistance pathways, CSC lineage tracing, and relapse modeling | Technical difficulty in isolation; risk of phenotypic drift during expansion | [96,97] |
| iPSC-Derived Tumor Organoids | Induced pluripotent stem cells (iPSCs) reprogrammed from patient somatic cells with engineered oncogenic alterations | Synthetic, chemically defined, or tunable hydrogels | Lineage-specific differentiation media coupled with CRISPR-based oncogenic mutation induction | High; amenable to automation and pooled genetic screens | Pediatric tumors, gliomas, colorectal, and pancreatic cancers | Modeling mutation-specific tumorigenesis, synthetic lethality studies, and organoid-based CRISPR screening | Complex and time-intensive differentiation protocols; potential for incomplete recapitulation of tumor heterogeneity | [78,98] |
| ESC-Derived Organoids | Pluripotent embryonic stem cells derived pre-implantation | Defined synthetic matrices or Matrigel | Developmental stage-specific media containing BMP4, FGF2, Activin A to induce tumor-relevant lineages | Low to moderate; ethical and regulatory limitations | Congenital tumors (hepatoblastoma, medulloblastoma, neuroblastoma) | Investigation of developmental origins of childhood tumors, modeling early oncogenic events | Restricted availability due to ethical constraints; limited clinical relevance for adult cancers | [99,100] |
| Progenitor-Derived Organoids | Pre-malignant progenitors from dysplastic or inflamed tissues | Collagen I or natural ECM-based scaffolds | Media mimicking inflammatory or pre-neoplastic microenvironments | Moderate; dependent on availability of early lesions | Barrett’s esophagus, IBD-associated CRC, gastric intestinal metaplasia | Modeling progression from inflammation to malignancy; studying chromosomal instability (CIN) and field cancerization | Limited to pre-invasive disease; often not reflective of invasive carcinoma biology | [101,102] |
| CTC-Derived Organoids | Circulating tumor cells isolated from patient blood samples | Hybrid systems combining ECM embedding with microfluidic capture or hanging-drop spheroids | Minimal survival-supporting media with ROCK inhibitor (ROCKi), B27 supplement, and antioxidants | Low; hindered by scarcity and fragility of CTCs | Metastatic breast, prostate, and non-small cell lung cancer (NSCLC) | Liquid biopsy-based real-time modeling of metastatic progression and treatment resistance evolution | Low cell yield; high culture failure rates; genetic drift possible during expansion | [103,104] |
| Model Type | Tumor Fidelity | Scalability | Microenvironmental Integration | Translational Utility | Emerging Innovations | Key Limitations | Source |
|---|---|---|---|---|---|---|---|
| 2D Cell Lines | Low, artificial monolayer growth, loss of heterogeneity | Very high, cost-efficient, and automation-compatible | Absent; lacks ECM, immune, or stromal signals | Routine molecular biology and initial drug screens | AI-driven image-based functional screening (e.g., Cell Painting, DeepCell), Lineage barcoding, and synthetic lethal drug screening | Lack of clinical correlation; poorly modeled in vivo responses | [106,107,108,109,121] |
| 3D Spheroids | Moderate; partial 3D cell–cell interaction, but clonal origin | Moderate; suitable for rapid testing | Limited; lacks immune or stromal cell types | Studies on drug penetration, metabolic gradients | Real-time metabolic imaging via biosensor dyes, used in combination with perfused scaffolds for dynamic testing | Lacks patient specificity and tissue context | [111,112,113,122] |
| Patient-Derived Xenografts (PDXs) | High; preserves tumor histology, subtype, and molecular profile | Low; requires months to establish and propagate | Native murine microenvironment; no functional human immunity unless humanized | In vivo efficacy, biomarker validation, and co-clinical trials | Humanized PDXs for immune checkpoint modeling, PDX transcriptomic–proteomic atlases (PDXNet, EurOPDX) | Costly, low throughput; species mismatch; limited to select cancer types | [5,115,116,117,118] |
| Organoids (PDOs) | High; retains tumor architecture, genetic identity, and intratumoral heterogeneity | Moderate to high; suitable for biobanking and automation | Partial; improved with TME co-cultures and microfluidics | Functional precision oncology, resistance profiling, and real-time clinical matching | AI-integrated phenotypic drug response prediction, Single-cell sequencing + spatial omics overlays, Organoid-on-chip for vasculature and flow simulation | Still lacks systemic inputs (e.g., circulation, hormones); needs standardization | [119,120,123] |
| Organotypic Tumor Slices (Live Tissue Cultures) | Very high; native tumor microenvironment retained for the short term | Low; viable for 5–10 days post-excision | Full microenvironment (fibroblasts, vessels, immune cells) intact | Short-term ex vivo drug response studies; immune profiling | Used for multiplexed immunotherapy testing (e.g., PD-1, TIL dynamics), Time-lapse imaging for tumor–immune interaction analysis | Limited lifespan, not scalable, variable reproducibility | [124,125] |
| Liquid Biopsy-Integrated Models (CTC/DNA-Coupled Organoids) | Patient-specific; reflects real-time mutational status | Low (nascent field); limited sample material | Minimal unless co-cultured; under development | Longitudinal tracking of resistance evolution; real-time personalization | CTC-derived PDOs for drug screening in metastatic settings, Cell-free DNA used to guide organoid mutation editing | Technically challenging, low cell recovery; requires enrichment tools | [57,126] |
| Co-Culture Type | Integrated Cell Types | Matrix or Platform Used | Experimental Applications | Key Functional Insights | Reference |
|---|---|---|---|---|---|
| Organoid–CAF Co-Culture | Tumor organoids + Cancer-associated fibroblasts (CAFs) | Matrigel, collagen I, or PEG-based synthetic ECM | ECM remodeling, tumor invasion, TME-driven resistance | Fibroblast-secreted factors (IL-6, VEGF-A); enhanced invasion; EMT; stromal-mediated drug resistance | [186,187] |
| Organoid–Endothelial Cell Co-Culture | Tumor organoids + HUVECs/EPCs | Fibrin or hyaluronic acid hydrogels; microfluidic chip | Modeling angiogenesis, perfusable vasculature | Vessel formation, endothelial barrier function, response to anti-angiogenics (e.g., bevacizumab) | [188,189] |
| Organoid–Immune Cell Co-Culture | Tumor organoids + PBMCs/TILs/NK cells | Air–liquid interface; transwell inserts; ALI chips | Immune activation, checkpoint response, cytotoxicity assays | T cell infiltration, IFN-γ secretion, PD-L1 upregulation, immune synapse formation, CAR-T specificity | [46,190] |
| Organoid–MSC Co-Culture | Tumor organoids + Mesenchymal stem/stromal cells | Dual-compartment ECM (Matrigel + collagen I) | Tumor–stromal crosstalk, cytokine delivery, immunomodulation | Cytokine gradients (TGF-β, IL-8), matrix stiffness modulation, and immune suppression modeling | [56,191] |
| Organoid–Neural Cell Co-Culture | Tumor organoids + Sensory ganglia or iPSC-derived neurons | Laminin-rich hydrogel, chip-based neural niches | Perineural invasion, neurotrophic signaling in tumors | Neurite extension, neurotransmitter effects (e.g., norepinephrine), and tumor migration toward nerve projections | [192,193] |
| Organoid–Dendritic Cell Co-Culture | Tumor organoids + moDCs or primary DCs | ECM dome + immune-compatible matrix (collagen IV) | Antigen presentation, neoantigen response prediction | DC maturation (CD83, CD86), T cell priming, neoantigen-specific responses, cytokine profiling | [194] |
| Multi-lineage Tumor–Immune–Stroma Chip | Organoids + fibroblasts + immune cells (T, NK, macrophages) | Perfused microfluidic chip with ECM scaffolding | Integrated TME simulation | Spatial phenotyping, immune evasion tracking, multi-cell interaction mapping, live-cell imaging | [195] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madan, A.; Saini, R.; Dhiman, N.; Juan, S.-H.; Satapathy, M.K. Organoids as Next-Generation Models for Tumor Heterogeneity, Personalized Therapy, and Cancer Research: Advancements, Applications, and Future Directions. Organoids 2025, 4, 23. https://doi.org/10.3390/organoids4040023
Madan A, Saini R, Dhiman N, Juan S-H, Satapathy MK. Organoids as Next-Generation Models for Tumor Heterogeneity, Personalized Therapy, and Cancer Research: Advancements, Applications, and Future Directions. Organoids. 2025; 4(4):23. https://doi.org/10.3390/organoids4040023
Chicago/Turabian StyleMadan, Ayush, Ramandeep Saini, Nainci Dhiman, Shu-Hui Juan, and Mantosh Kumar Satapathy. 2025. "Organoids as Next-Generation Models for Tumor Heterogeneity, Personalized Therapy, and Cancer Research: Advancements, Applications, and Future Directions" Organoids 4, no. 4: 23. https://doi.org/10.3390/organoids4040023
APA StyleMadan, A., Saini, R., Dhiman, N., Juan, S.-H., & Satapathy, M. K. (2025). Organoids as Next-Generation Models for Tumor Heterogeneity, Personalized Therapy, and Cancer Research: Advancements, Applications, and Future Directions. Organoids, 4(4), 23. https://doi.org/10.3390/organoids4040023









