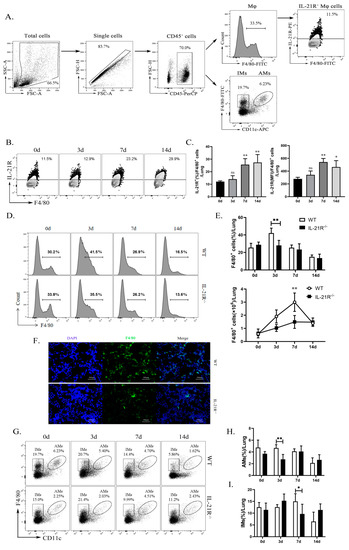
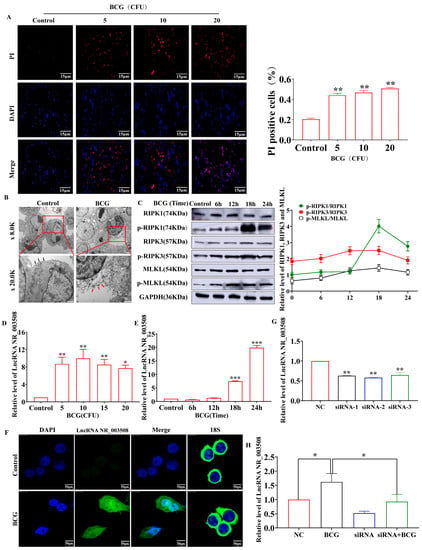
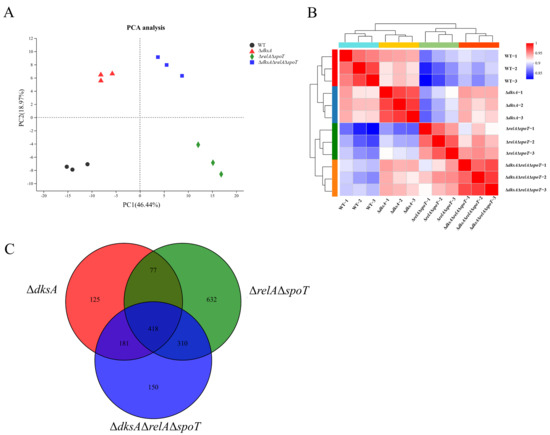
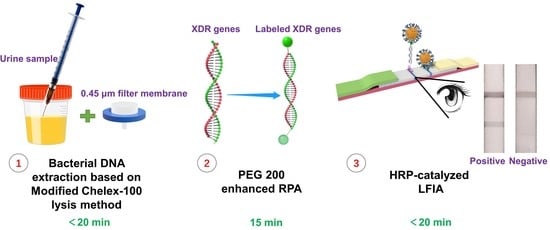
State-of-the-Art Molecular Microbiology in China
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Microbiology".
Submission Status: Closed | Viewed by 51082Editors
Interests: ruminal bacteria
Interests: wheat disease resistance; soil-borne fungual diseases; resistance-associated genes; comparative transcriptome; gene functional study
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topical Collection aims to provide a comprehensive overview of recent advances in molecular microbiology in China by inviting contributions from Chinese research institutes/laboratories that consolidate our understanding of this area. Topics include, but are not limited to, the following:
- Antibiotic resistance mechanisms;
- Biosynthesis of macromolecules;
- Cell division and cell wall structure;
- Gene expression and its regulation;
- Gene transfer mechanisms;
- Host–pathogen interactions, including host responses;
- Induction of cell death by microorganisms;
- Membrane biogenesis and function;
- Pathogenicity mechanisms;
- Post-translational modifications;
- Protein delivery (secretion and trafficking);
- Signaling pathways and networks;
- Systems biology;
- Vaccines;
- Virulence factors;
- Antimicrobial peptides;
- Therapeutic strategies;
- Nutrients metabolism;
- Genomcis and metagenomics;
- Interaction of microbes with host;
- Protein structure and function;
- Probiotics;
- Communication of microbes;
- Synthetic biology;
- Animal and human gut health.
Dr. Shengguo Zhao
Dr. Zengyan Zhang
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.