LncRNA NR_003508 Suppresses Mycobacterium tuberculosis-Induced Programmed Necrosis via Sponging miR-346-3p to Regulate RIPK1
Abstract
1. Introduction
2. Results
2.1. BCG-Infection Stimulates Cell-Programmed Necrosis Accompanies with Increased LncRNA NR_003508 Expression
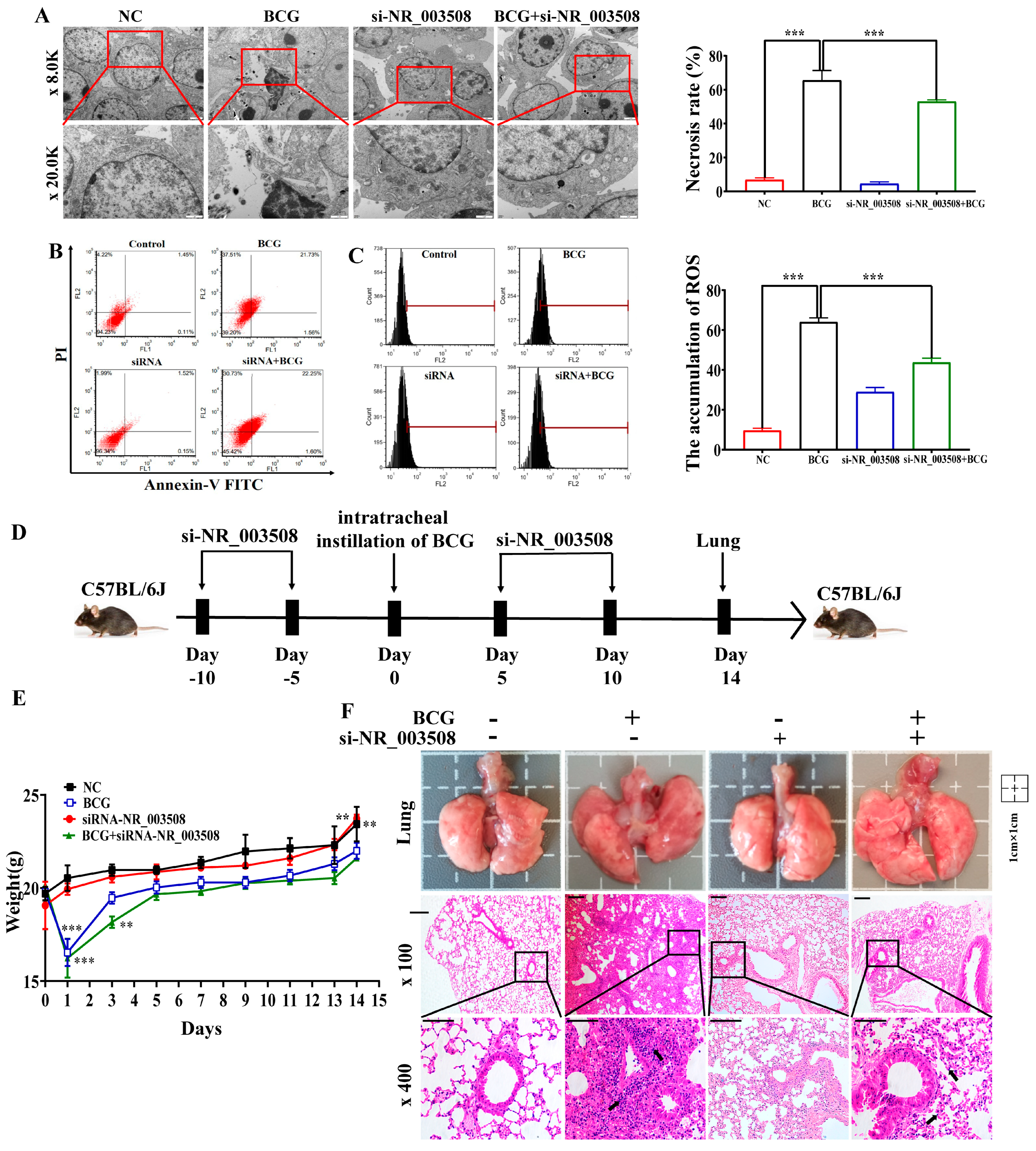
2.2. si-NR_003508 Attenuates Mycobacterium Tuberculosis Induced Programmed Necrosis In Vitro and In Vivo
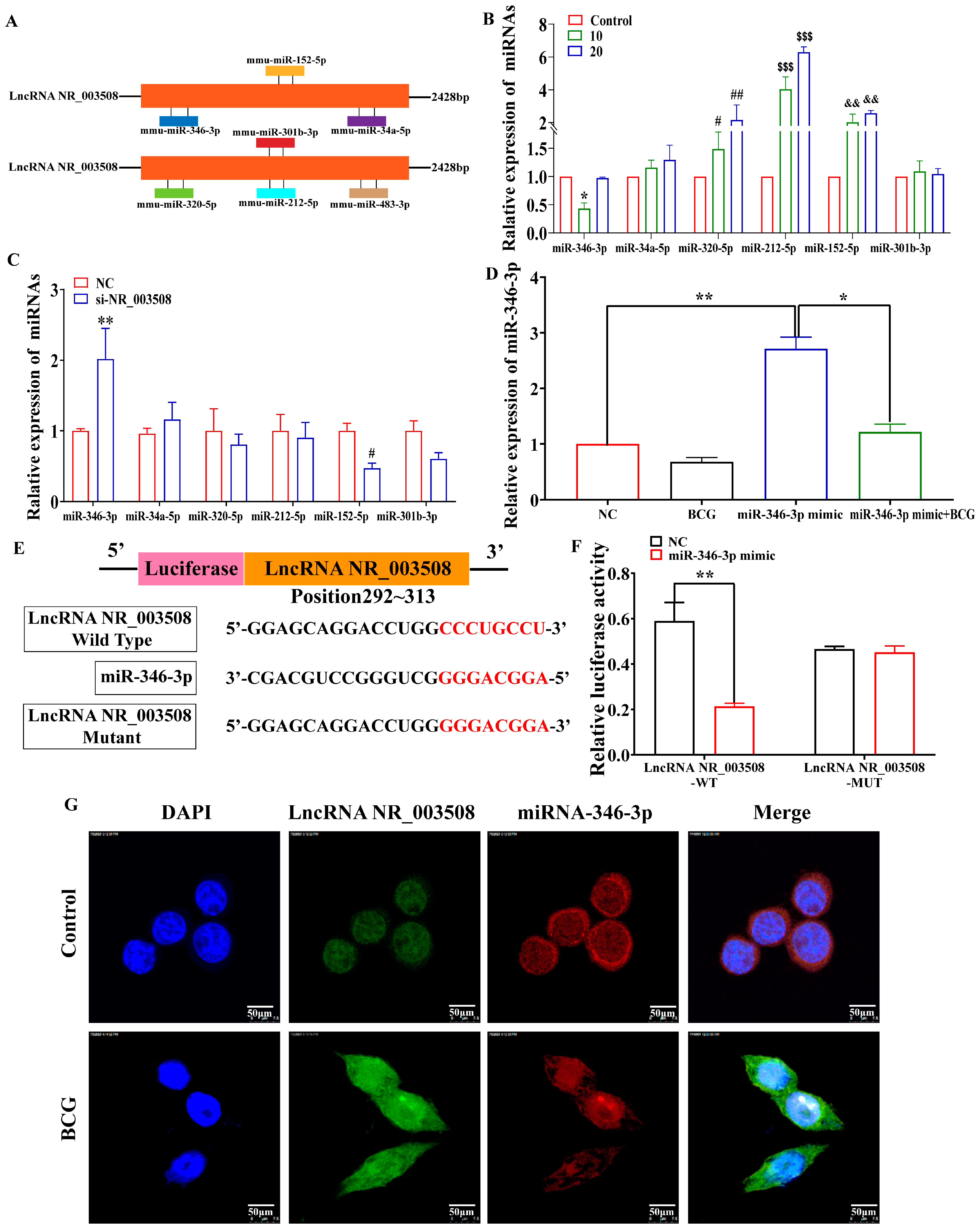
2.3. LncRNA NR_003508 Functions as a Competing Endogenous RNA by Binding to miR-346-3p
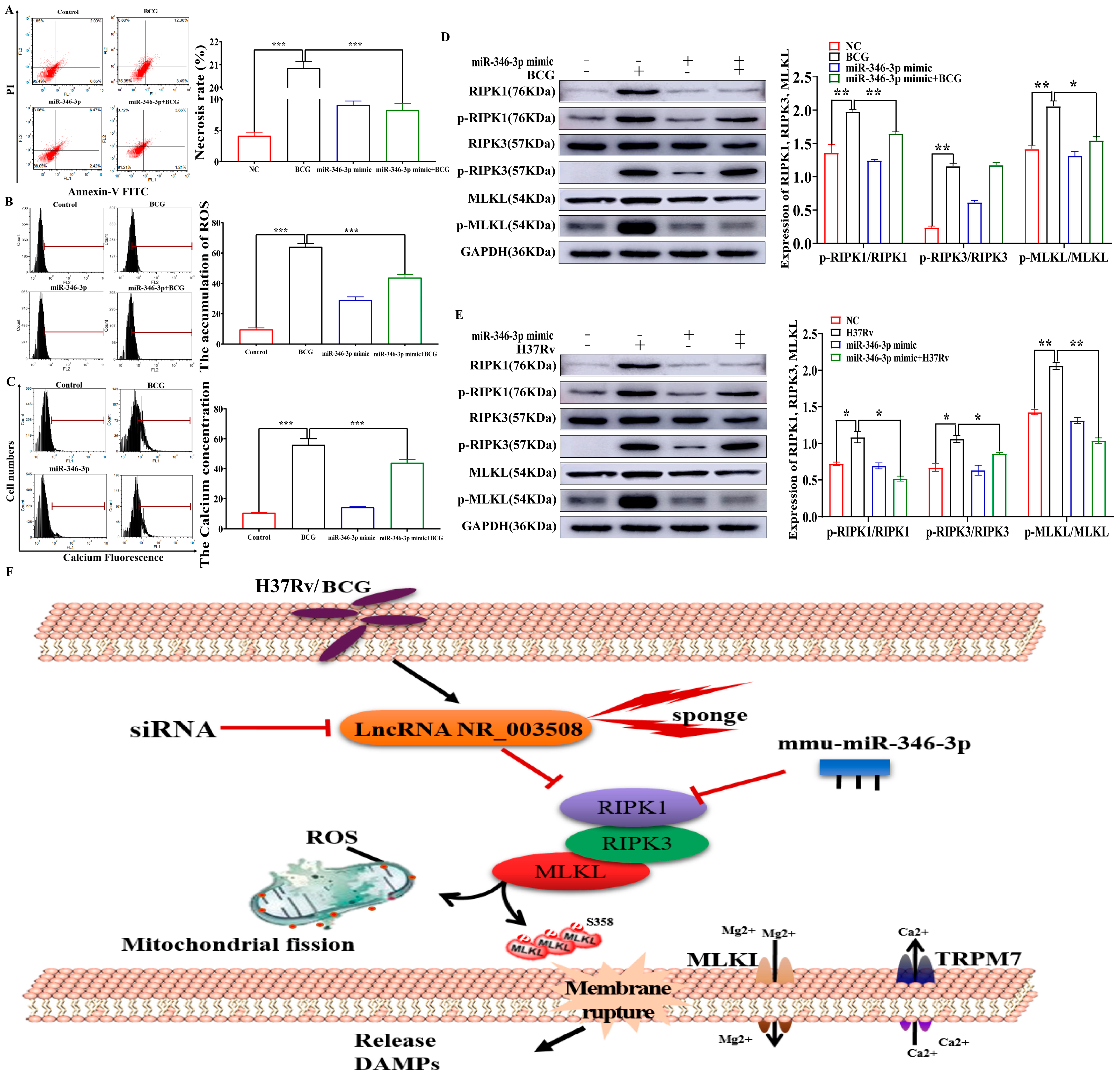
2.4. miR-346-3p Targets RIPK1 to Regulate Programmed Necrosis
2.5. miR-346-3p Participates in Programmed Necrosis through RIPK1/RIPK3/MLKL Axis
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Bacterial Strain
4.2. Clinical Sample Collection
4.3. Ethics and Animals
4.4. Transfection of Small Interfering RNA (siRNA) and Plasmid
4.5. Everse Transcription-Polymerase Chain Reaction (qRT-PCR)
4.6. Western Blot
4.7. Immunofluorescence Assay
4.8. PI Staining and Confocal Microscope Assay for Cell Necrosis
4.9. Fluorescence in Situ Hybridization (FISH)
4.10. Annexin V and PI Staining for Apoptosis and Necrosis by Flow Cytometry
4.11. Determination of the ROS and Ca2+ Level
4.12. Luciferase Reporter Assay
4.13. Hematoxylin-Eosin (H&E) Staining
4.14. Transmission Electron Microscopy (TEM) Analysis
4.15. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schito, M.; Migliori, G.B.; Fletcher, H.A.; McNerney, R.; Centis, R.; D’Ambrosio, L.; Bates, M.; Kibiki, G.; Kapata, N.; Corrah, T.; et al. Perspectives on Advances in Tuberculosis Diagnostics, Drugs, and Vaccines. Clin. Infect. Dis. 2015, 61 (Suppl. 3), S102–S118. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Tuberculosis Report 2021; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Shariq, M.; Quadir, N.; Sharma, N.; Singh, J.; Sheikh, J.A.; Khubaib, M.; Hasnain, S.E.; Ehtesham, N.Z. Mycobacterium tuberculosis RipA Dampens TLR4-Mediated Host Protective Response Using a Multi-Pronged Approach Involving Autophagy, Apoptosis, Metabolic Repurposing, and Immune Modulation. Front. Immunol. 2021, 12, 636644. [Google Scholar] [CrossRef]
- Liu, G.M.; Wan, Q.F.; Li, J.W.; Hu, X.Y.; Gu, X.L.; Xu, S.C. Silencing miR-125b-5p attenuates inflammatory response and apoptosis inhibition in mycobacterium tuberculosis-infected human macrophages by targeting DNA damage-regulated autophagy modulator 2 (DRAM2). Cell Cycle 2020, 19, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Mitini-Nkhoma, S.C.; Chimbayo, E.T.; Mzinza, D.T.; Mhango, D.V.; Chirambo, A.P.; Mandalasi, C.; Lakudzala, A.E.; Tembo, D.L.; Jambo, K.C.; Mwandumba, H.C. Something Old, Something New: Ion Channel Blockers as Potential Anti-Tuberculosis Agents. Front. Immunol. 2021, 12, 665785. [Google Scholar] [CrossRef]
- Min, W.L.; Sun, L.Z.; Li, B.R.; Gao, X.; Zhang, S.Q.; Zhao, Y. lncCRLA Enhanced Chemoresistance in Lung Adenocarcinoma That Underwent EpithelialMesenchymal Transition. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2022, 28, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Pal, S.; Garg, M.; Pandey, A.K. Deciphering the Mounting Complexity of the p53 Regulatory Network in Correlation to Long Non-Coding RNAs (lncRNAs) in Ovarian Cancer. Cells 2020, 9, 527. [Google Scholar] [CrossRef]
- Li, G.; Liu, K.; Du, X. Long Non-Coding RNA TUG1 Promotes Proliferation and Inhibits Apoptosis of Osteosarcoma Cells by Sponging miR-132-3p and Upregulating SOX4 Expression. Yonsei Med. J. 2018, 59, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C.; Su, Z.L.; Lu, S.N.; Fu, W.; Liu, Z.F.; Jiang, X.M.; Tai, S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin. Chim. Acta 2018, 485, 229–233. [Google Scholar] [CrossRef]
- Yang, Z.; Jiang, S.A.; Shang, J.J.; Jiang, Y.T.; Dai, Y.; Xu, B.P.; Yu, Y.; Liang, Z.X.; Yang, Y. LncRNA: Shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res. Rev. 2019, 52, 17–31. [Google Scholar] [CrossRef]
- Tao, H.; Liu, X.P.; Liu, X.X.; Liu, W.F.; Wu, D.; Wang, R.L.; Lv, G. LncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling. Mol. Reprod. Dev. 2019, 86, 686–695. [Google Scholar] [CrossRef]
- Wang, K.; Liu, F.; Liu, C.Y.; An, T.; Zhang, J.; Zhou, L.Y.; Wang, M.; Dong, Y.H.; Li, N.; Gao, J.N.; et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016, 23, 1394–1405. [Google Scholar] [CrossRef] [PubMed]
- Afriyie-Asante, A.; Dabla, A.; Dagenais, A.; Berton, S.; Smyth, R.; Sun, J. Mycobacterium tuberculosis Exploits Focal Adhesion Kinase to Induce Necrotic Cell Death and Inhibit Reactive Oxygen Species Production. Front. Immunol. 2021, 12, 742370. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Khan, N.; Gan, H.; Tzelepis, F.; Nishimura, T.; Park, S.Y.; Divangahi, M.; Remold, H.G. Bcl-xL mediates RIPK3-dependent necrosis in M. tuberculosis-infected macrophages. Mucosal Immunol. 2017, 10, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Zhang, X.J.; Li, Q.; Wang, K.; Wang, Y.; Jiao, J.Q.; Feng, C.; Teng, S.; Zhou, L.Y.; Gong, Y.; et al. MicroRNA-103/107 Regulate Programmed Necrosis and Myocardial Ischemia/Reperfusion Injury Through Targeting FADD. Circ. Res. 2015, 117, 352–363. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.L.; Ma, C.J.; Ma, Q.M.; Zhuang, P.P.; Deng, G.C. Microarray Profiling and Co-Expression Network Analysis of LncRNAs and mRNAs in Acute Respiratory Distress Syndrome Mouse Model. Pathogens 2022, 11, 532. [Google Scholar] [CrossRef]
- Sun, Q.; Shen, X.; Wang, P.; Ma, J.; Sha, W. Targeting cyclophilin-D by miR-1281 protects human macrophages from Mycobacterium tuberculosis-induced programmed necrosis and apoptosis. Aging 2019, 11, 12661–12673. [Google Scholar] [CrossRef]
- Fan, Y.; Sheng, W.W.; Meng, Y.; Cao, Y.D.; Li, R. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif. Cells Nanomed. Biotechnol. 2020, 48, 393–407. [Google Scholar] [CrossRef]
- Ni, C.; Fang, Q.Q.; Chen, W.Z.; Jiang, J.X.; Jiang, Z.; Ye, J.; Zhang, T.; Yang, L.; Meng, F.B.; Xia, W.J.; et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+gammadelta1 Treg cells. Signal Transduct. Target. Ther. 2020, 5, 41. [Google Scholar] [CrossRef]
- Piao, X.; Byun, H.S.; Lee, S.R.; Ju, E.J.; Park, K.A.; Sohn, K.C.; Quan, K.T.; Lee, J.; Na, M.; Hur, G.M. 8-Geranylumbelliferone isolated from Paramignya trimera triggers RIPK1/RIPK3-dependent programmed cell death upon TNFR1 ligation. Biochem. Pharmacol. 2021, 192, 114733. [Google Scholar] [CrossRef]
- Cambier, C.; Falkow, S.; Ramakrishnan, L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 2014, 159, 1497–1509. [Google Scholar] [CrossRef]
- Liu, C.H.; Liu, H.; Ge, B. Innate immunity in tuberculosis: Host defense vs pathogen evasion. Cell. Mol. Immunol. 2017, 14, 963–975. [Google Scholar] [PubMed]
- Wang, J.; Ge, P.P.; Lei, Z.H.; Lu, Z.; Qiang, L.H.; Chai, Q.Y.; Zhang, Y.; Zhao, D.D.; Li, B.X.; Su, J.Q.; et al. Mycobacterium tuberculosis protein kinase G acts as an unusual ubiquitinating enzyme to impair host immunity. EMBO Rep. 2021, 22, e52175. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, G.; Shrivastva, R.; Mukhopadhyay, S. Mycobacterial PknG Targets the Rab7l1 Signaling Pathway To Inhibit Phagosome-Lysosome Fusion. J. Immunol. 2018, 201, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chen, J.X.; Wang, P.; Li, H.H.; Zhou, Y.L.; Liu, H.P.; Liu, Z.H.; Zheng, R.J.; Wang, L.; Yang, H.; et al. MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat. Commun. 2018, 9, 4295. [Google Scholar] [CrossRef] [PubMed]
- Hua, Q.; Jin, M.M.; Mi, B.M.; Xu, F.; Li, T.; Zhao, L.; Liu, J.J.; Huang, G. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J. Hematol. Oncol. 2019, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.Y.; Chen, X.L.; He, Y.T.; Xue, C.; Ren, F.; Ren, Z.G.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef]
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef]
- Tian, F.; Wang, J.H.; Zhang, Z.H.; Yang, J. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol. Res. 2020, 53, 9. [Google Scholar] [CrossRef]
- Jianfang, W.; Hui, W.; Le, K. LINC00870 regulates Th1/Th2 via the JAK/STAT pathway in peripheral blood mononuclear cells infected with Mycobacterium tuberculosis. Int. Immunopharmacol. 2022, 102, 107188. [Google Scholar] [CrossRef]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.W.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Shan, H.J.; Zhang, P.; She, C.; Zhou, X.Z. LncRNA EPIC1 protects human osteoblasts from dexamethasone-induced cell death. Biochem. Biophys. Res. Commun. 2018, 503, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, F.; Xu, F.Y.; Wang, Q.; Wu, J.H.; Peng, W.X.; Dong, W.T. LncRNA NEAT1 regulates CYP1A2 and influences steroid-induced necrosis. Open Life Sci. 2021, 16, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Gu, X.Y.; Li, G.L.; Bao, Z.Y.; Li, L.J. Mitochondrial Mechanisms of Necroptosis in Liver Diseases. Int. J. Mol. Sci. 2020, 22, 66. [Google Scholar] [CrossRef] [PubMed]
- Sargazi, S.; Mukhtar, M.; Rahdar, A.; Bilal, M.; Barani, M.; Díez-Pascual, A.M.; Behzadmehr, R.; Pandey, S. Opportunities and challenges of using high-sensitivity nanobiosensors to detect long noncoding RNAs: A preliminary review. Int. J. Biol. Macromol. 2022, 205, 304–315. [Google Scholar] [CrossRef]
- Quan, M.; Chen, J.; Zhang, D. Exploring the secrets of long noncoding RNAs. Int. J. Mol. Sci. 2015, 16, 5467–5496. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Wang, G.Y.; Liang, X.L.; Wu, Z.; Wang, X.C.; Dong, Z.M.; Guo, Y.L.; Shen, S.P.; Liang, J.; Guo, W. LncRNA FAM83H-AS1 promotes oesophageal squamous cell carcinoma progression via miR-10a-5p/Girdin axis. J. Cell. Mol. Med. 2020, 24, 8962–8976. [Google Scholar] [CrossRef]
- Liu, S.; Yin, H.H.; Zheng, S.W.; Chu, A.N.; Li, Y.Z.; Xing, C.Z.; Yuan, Y.; Gong, Y.H. Differentially Expressed mRNAs and Their Long Noncoding RNA Regulatory Network with Helicobacter pylori-Associated Diseases including Atrophic Gastritis and Gastric Cancer. BioMed Res. Int. 2020, 2020, 3012193. [Google Scholar] [CrossRef]
- Li, Z.; Cai, B.L.; Abdalla, B.A.; Zhu, X.N.; Zheng, M.; Han, P.G.; Nie, Q.H.; Zhang, X.Q. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J. Cachexia Sarcopenia Muscle 2019, 10, 391–410. [Google Scholar] [CrossRef]
- Wang, M.; Mao, C.; Ouyang, L.L.; Liu, Y.T.; Lai, W.W.; Liu, N.; Shi, Y.; Chen, L.; Xiao, D.S.; Yu, F.L.; et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019, 26, 2329–2343. [Google Scholar] [CrossRef]
- Yin, J.; Zeng, X.L.; Ai, Z.X.; Yu, M.; Wu, Y.O.; Li, S.J. Construction and analysis of a lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in oral cancer. BMC Med. Genom. 2020, 13, 84. [Google Scholar] [CrossRef]
- Cao, W.; Zhou, G. LncRNA SNHG12 contributes proliferation, invasion and epithelial-mesenchymal transition of pancreatic cancer cells by absorbing miRNA-320b. Biosci. Rep. 2020, 40, BSR20200805. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Lou, J.; Zheng, X.M.; Yang, X.Y. LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by Mycobacterium tuberculosis through the miR-665/ULK1 signaling axis. Mol. Immunol. 2021, 139, 42–49. [Google Scholar] [CrossRef]
- Miliotis, C.; Slack, F.J. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021, 518, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, K.X.; Zhou, H.M.; Jiang, L.; Xie, B.; Wang, R.Y.; Xia, W.Z.; Yin, Y.J.; Gao, Z.Y.; Cui, D.S.; et al. Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer’s Disease. Aging Cell 2020, 19, e13125. [Google Scholar] [CrossRef] [PubMed]
- Avsar, B.; Zhao, Y.H.; Li, W.H.; Lukiw, W.J. Atropa belladonna Expresses a microRNA (aba-miRNA-9497) Highly Homologous to Homo sapiens miRNA-378 (hsa-miRNA-378); both miRNAs target the 3’-Untranslated Region (3’-UTR) of the mRNA Encoding the Neurologically Relevant, Zinc-Finger Transcription Factor ZNF-691. Cell. Mol. Neurobiol. 2020, 40, 179–188. [Google Scholar] [PubMed]
- Lin, C.W.; Jan, M.S.; Kuo, J.H.S. Exploring MicroRNA Expression Profiles Related to the mTOR Signaling Pathway in Mouse Embryonic Fibroblast Cells Treated with Polyethylenimine. Mol. Pharm. 2015, 12, 2858–2868. [Google Scholar] [CrossRef]
- Li, J.; McQuade, T.; Siemer, A.B.; Napetschnig, J.; Moriwaki, K.; Hsiao, Y.S.; Damko, E.; Moquin, D.; Walz, T.; McDermott, A.; et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 2012, 150, 339–350. [Google Scholar] [CrossRef]
- Li, L.; Tong, A.; Zhang, Q.S.; Wei, Y.Q.; Wei, X.W. The molecular mechanisms of MLKL-dependent and MLKL-independent necrosis. J. Mol. Cell Biol. 2021, 13, 3–14. [Google Scholar] [CrossRef]
- Amaral, E.P.; Foreman, T.W.; Namasivayam, S.; Hilligan, K.L.; Kauffman, K.D.; Bomfim, C.C.B.; Costa, D.L.; Barreto-Duarte, B.; Gurgel-Rocha, C.; Santana, M.F.; et al. GPX4 regulates cellular necrosis and host resistance in Mycobacterium tuberculosis infection. J. Exp. Med. 2022, 219, e20220504. [Google Scholar] [CrossRef]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar]
- Sun, C.B.; Fu, Y.H.; Gu, X.; Xi, X.W.; Peng, X.; Wang, C.H.; Sun, Q.; Wang, X.Y.; Qian, F.C.; Qin, Z.F.; et al. Macrophage-Enriched lncRNA RAPIA: A Novel Therapeutic Target for Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1464–1478. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, L.; Shang, K.; Liu, F.; Che, J.J.; Li, H.H.; Cao, B.W. Long non-coding RNA H19, a novel therapeutic target for pancreatic cancer. Mol. Med. 2020, 26, 30. [Google Scholar] [CrossRef] [PubMed]



| Sense (5′-3′) | Antisense (5′-3′) | |
|---|---|---|
| siRNA-1 | GCGUUGAUUCAGUCAACUUTT | AAGUUGACUGAAUCAACGCTT |
| siRNA-2 | GCACACGGUCACUGAAAUUTT | AAUUUCAGUGACCGUGUGCTT |
| siRNA-3 | ACACUGCUAGAAAUAAAUUTT | AAUUUAUUUCUAGCAGUGUGC |
| Primer | Sequence (5′-3′) |
|---|---|
| LncRNA NR_003508 | F: GTATGAGGAGAAGGTGCGGC |
| R: CCAGAACTCTGGTCCCCAAT | |
| RIPK1 | F: AGAACAACCTGGATCGCTGC |
| R: CCTGCACACTGCGATCATT | |
| RIPK3 | F: CCTTCAGAGGCACAACACCT |
| R: TGTCATTGGATTCGGTGGG | |
| MLKL | F GCCACTGGAAAGATCCCATTTG |
| R: TTCCCGCAACAACTCAGG | |
| GAPDH | F: TACCCCCAATGTGTCCGTC |
| R: AAGAGTGGGAGTTGCTGTTGAAG | |
| miR-346-3p | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTGCAGGC |
| F: ACACTCCAGCTGGCAGGCAGGGGCTGGGCCTG | |
| miR-34a-5p | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGCTGCAGGC |
| F: ACACTCCAGCTGGCCGGGAACGTCGAGACTGG | |
| miR-320-5p | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGGAAGAGA |
| F: ACACTCCAGCTGGCTTCCCTTTGTCATCCTTT | |
| miR-212-5p | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGAACCGA |
| F: ACACTCCAGCTGGCAGGGGCTGGCTTTCCTCT | |
| miR-152-5p | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCAAGACA |
| F: ACACTCCAGCTGGCAGAGGGTGCGCCGGGCCCT | |
| miR-301b-3p | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTCACGTTA |
| F: ACACTCCAGCTGGCAGGAAGCCCTGGAGGGGCT | |
| miRNA Universal Reverse sequence | TGGTGTCGTGGAGTCG |
| SnoRNA202 | RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCTGGAAT |
| F: ACACTCCAGCTGGCAGTCAGGCTCCTGGCTAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Yu, Z.; Ma, Q.; Yu, J.; Gong, Z.; Deng, G.; Wu, X. LncRNA NR_003508 Suppresses Mycobacterium tuberculosis-Induced Programmed Necrosis via Sponging miR-346-3p to Regulate RIPK1. Int. J. Mol. Sci. 2023, 24, 8016. https://doi.org/10.3390/ijms24098016
Liu L, Yu Z, Ma Q, Yu J, Gong Z, Deng G, Wu X. LncRNA NR_003508 Suppresses Mycobacterium tuberculosis-Induced Programmed Necrosis via Sponging miR-346-3p to Regulate RIPK1. International Journal of Molecular Sciences. 2023; 24(9):8016. https://doi.org/10.3390/ijms24098016
Chicago/Turabian StyleLiu, Li, Zhirui Yu, Qinmei Ma, Jialin Yu, Zhaoqian Gong, Guangcun Deng, and Xiaoling Wu. 2023. "LncRNA NR_003508 Suppresses Mycobacterium tuberculosis-Induced Programmed Necrosis via Sponging miR-346-3p to Regulate RIPK1" International Journal of Molecular Sciences 24, no. 9: 8016. https://doi.org/10.3390/ijms24098016
APA StyleLiu, L., Yu, Z., Ma, Q., Yu, J., Gong, Z., Deng, G., & Wu, X. (2023). LncRNA NR_003508 Suppresses Mycobacterium tuberculosis-Induced Programmed Necrosis via Sponging miR-346-3p to Regulate RIPK1. International Journal of Molecular Sciences, 24(9), 8016. https://doi.org/10.3390/ijms24098016






