Antifungal Substances Produced by Xenorhabdus bovienii and Its Inhibition Mechanism against Fusarium solani
Abstract
:1. Introduction
2. Results
2.1. Antifungal Substances in the Supernatant of X. bovienii Ferment
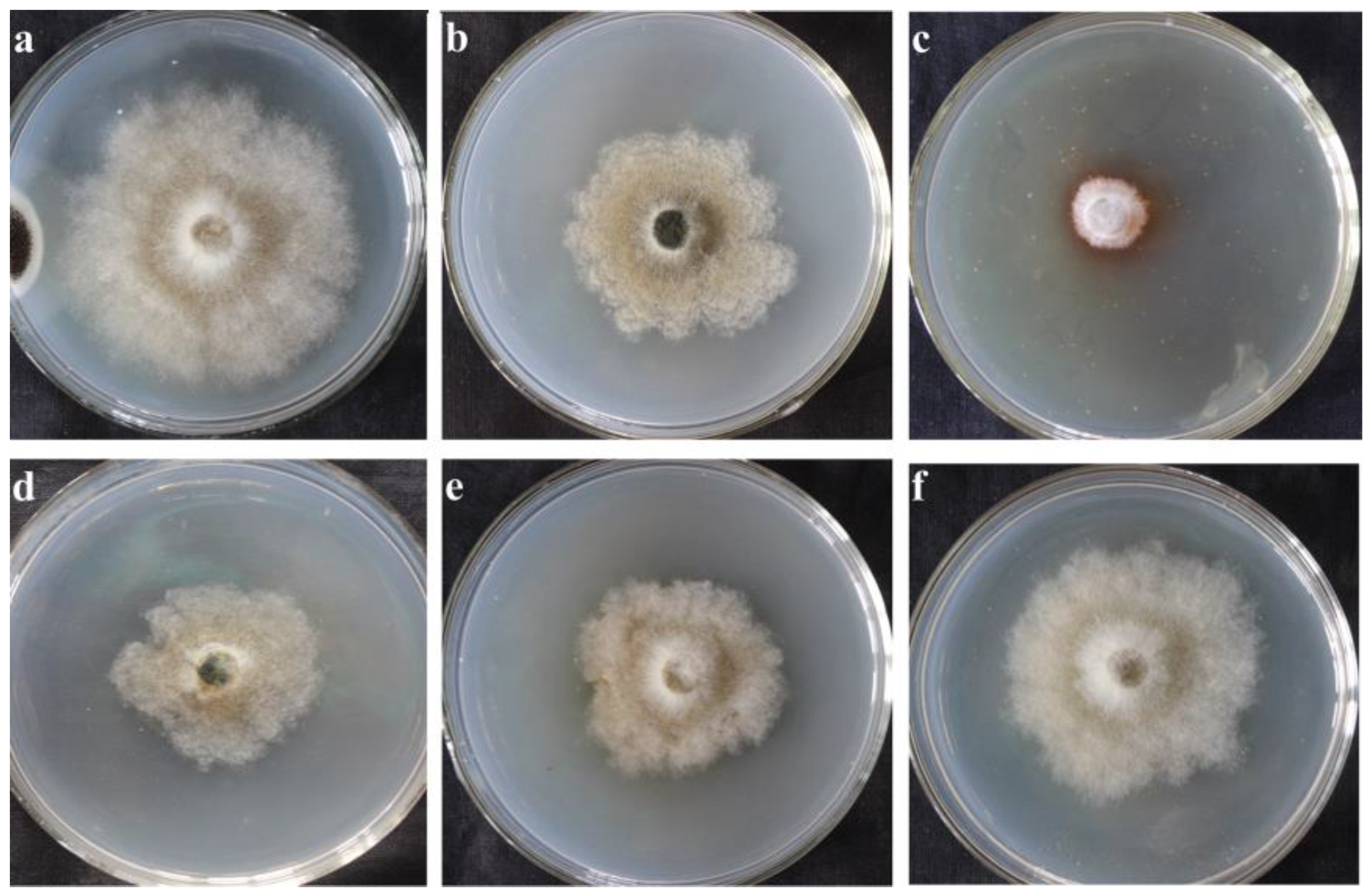
2.1.1. Inhibitory Effect of Incubation with the Supernatant of Fermented X. bovienii for Different Time
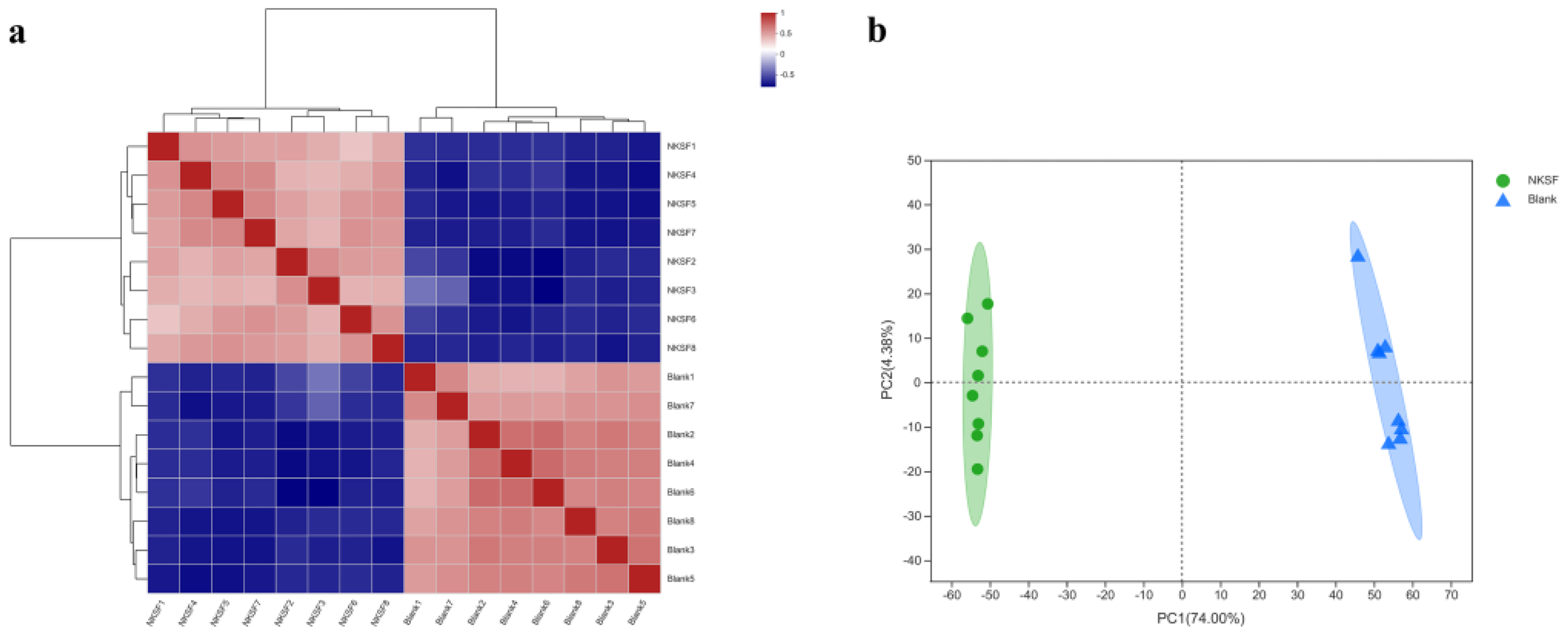
2.1.2. Correlation Analysis of Metabolomic Data
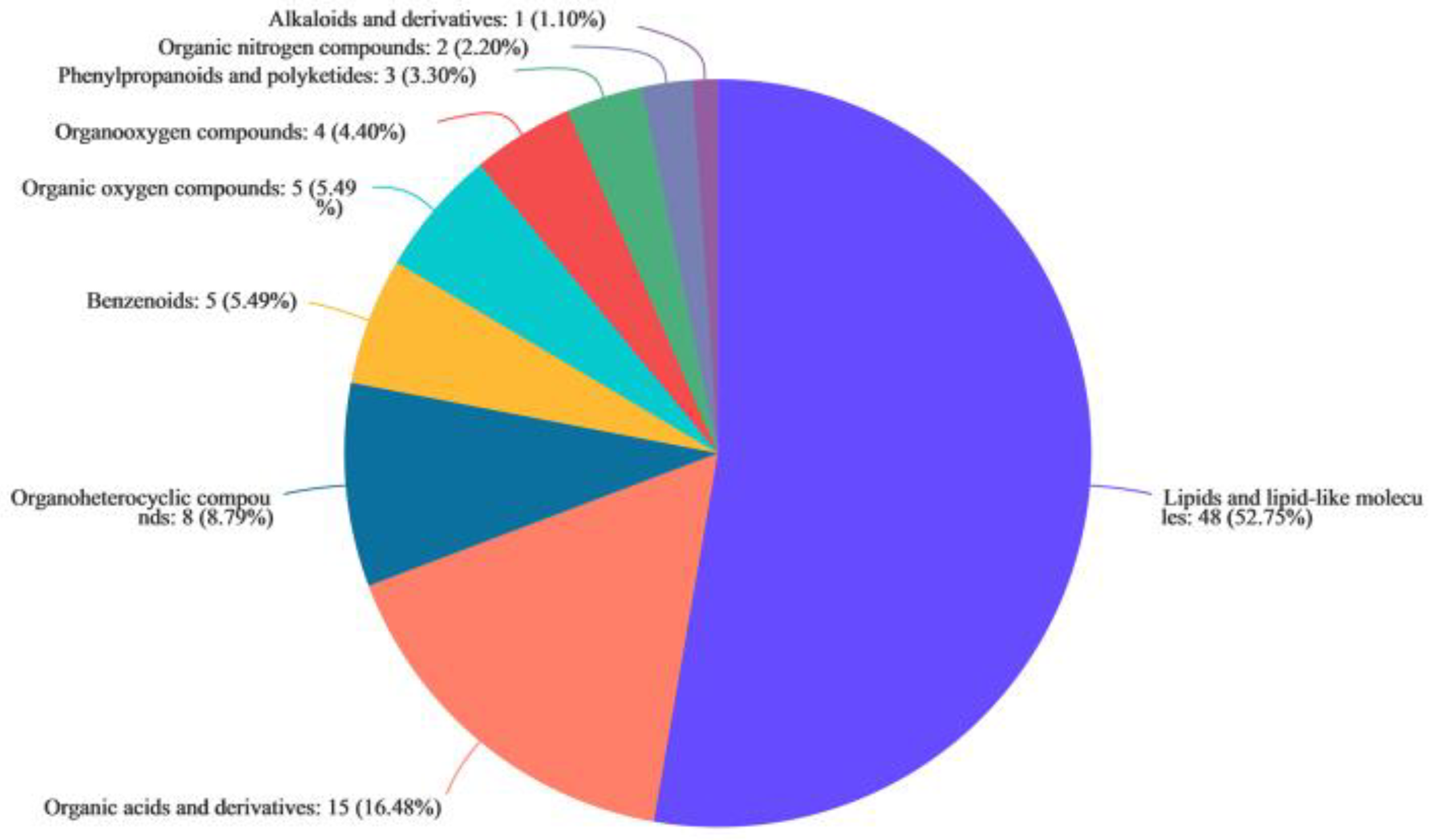
2.1.3. Metabolites in the Supernatant of X. bovienii Ferment
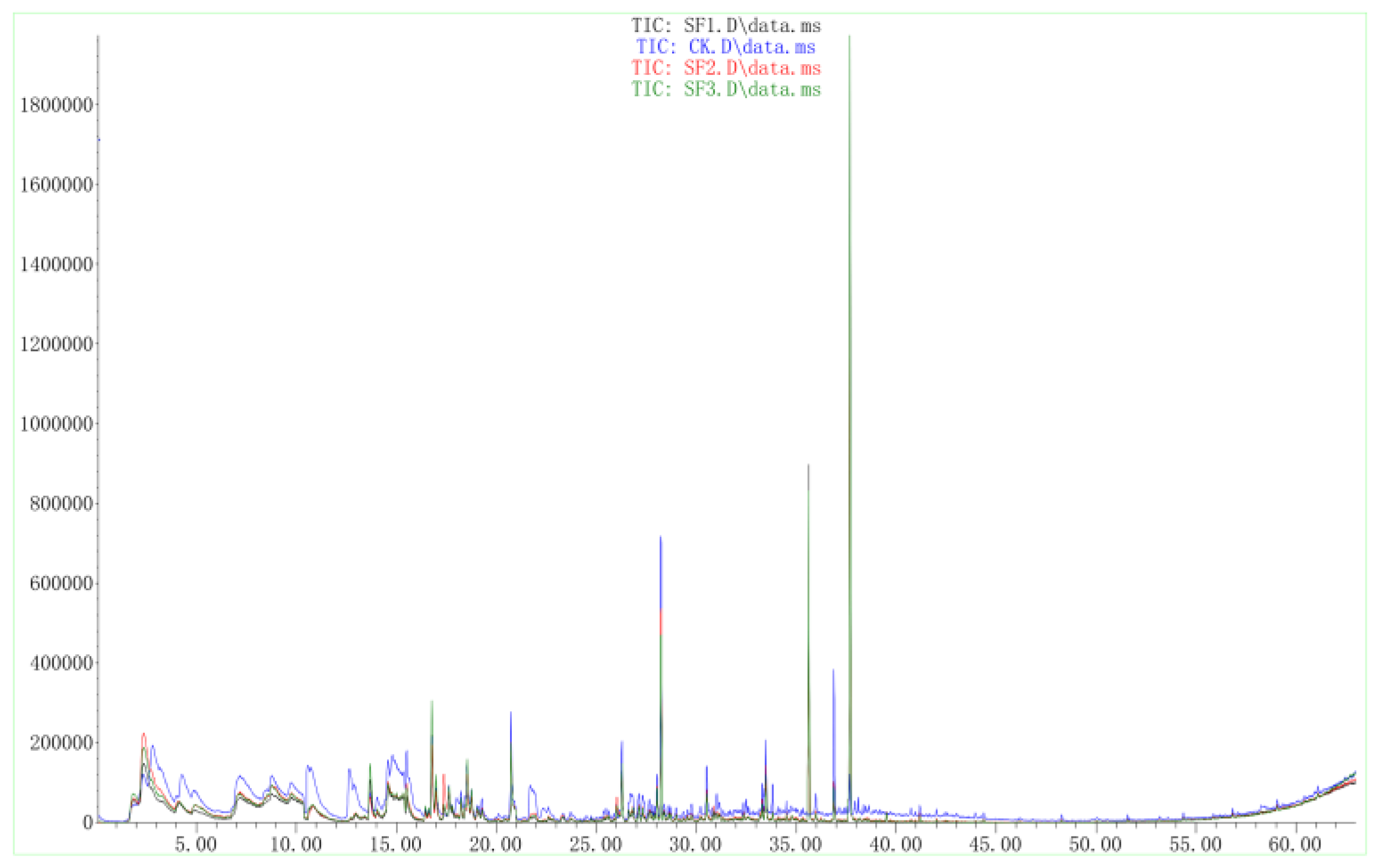
2.2. Volatile Antifungal Substances Produced by X. bovienii
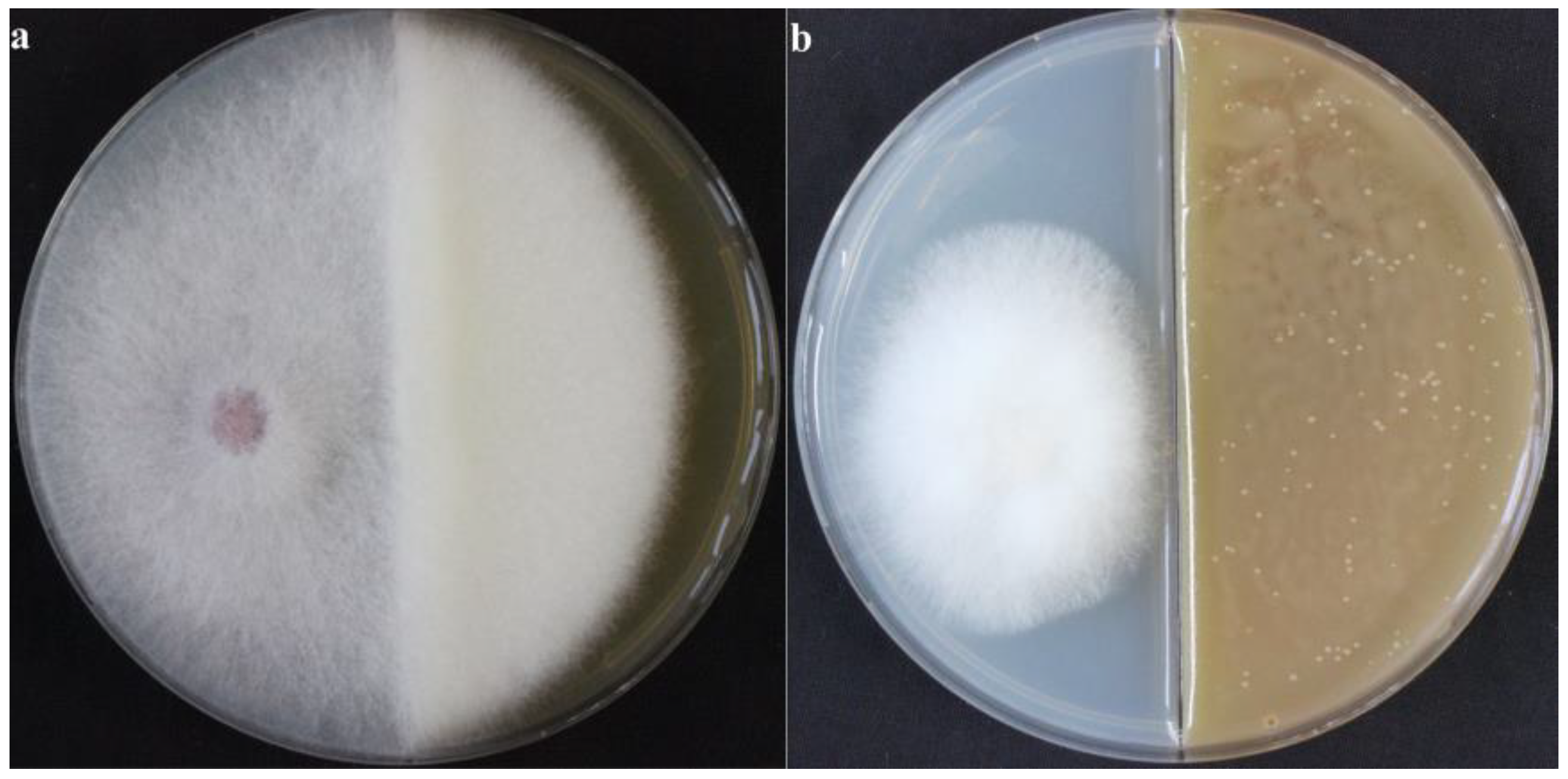
2.2.1. Inhibitory Effect of VOCs Produced by X. bovienii on NK-NH1
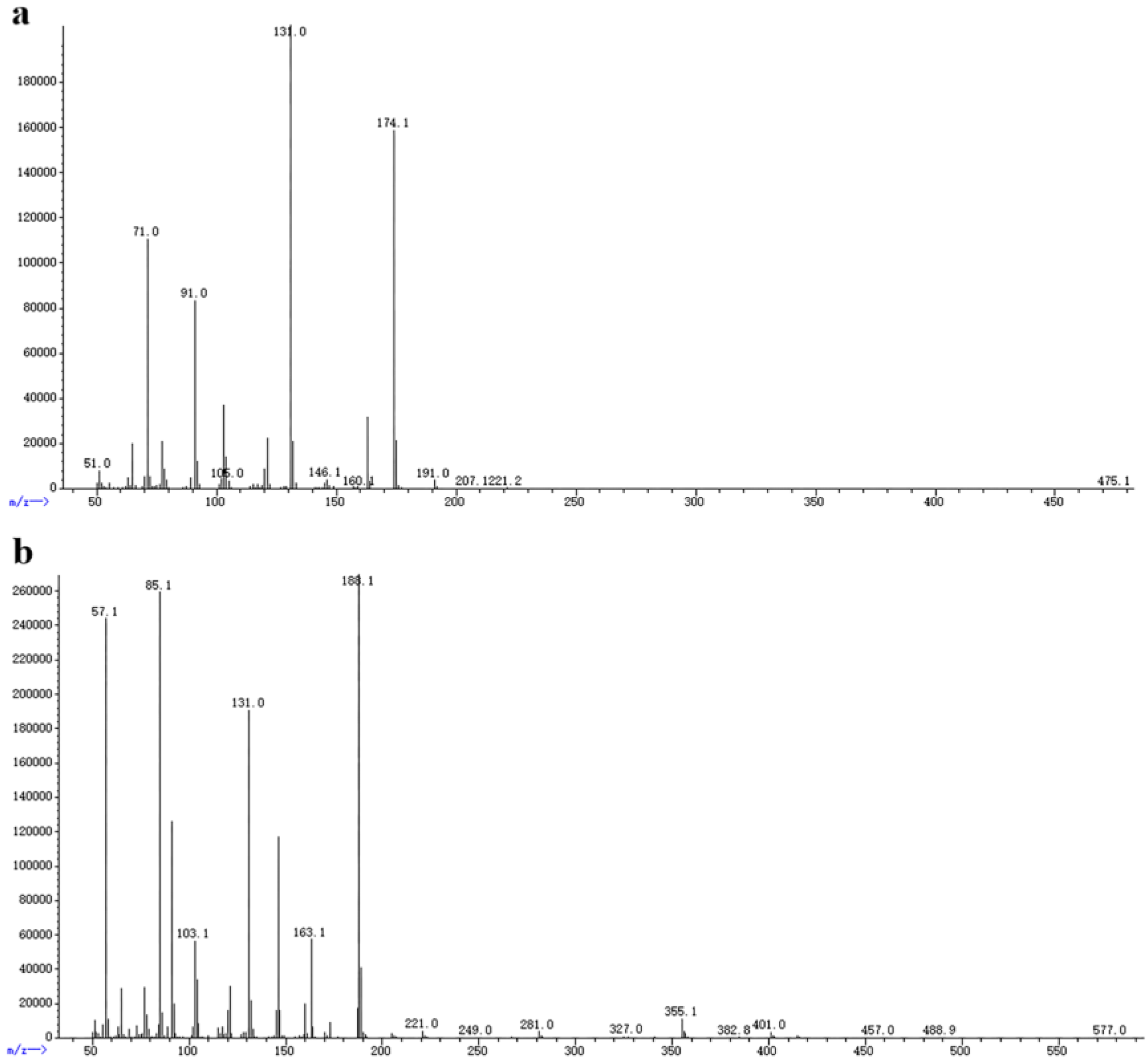
2.2.2. GC–MS of VOCs Produced by X. bovienii
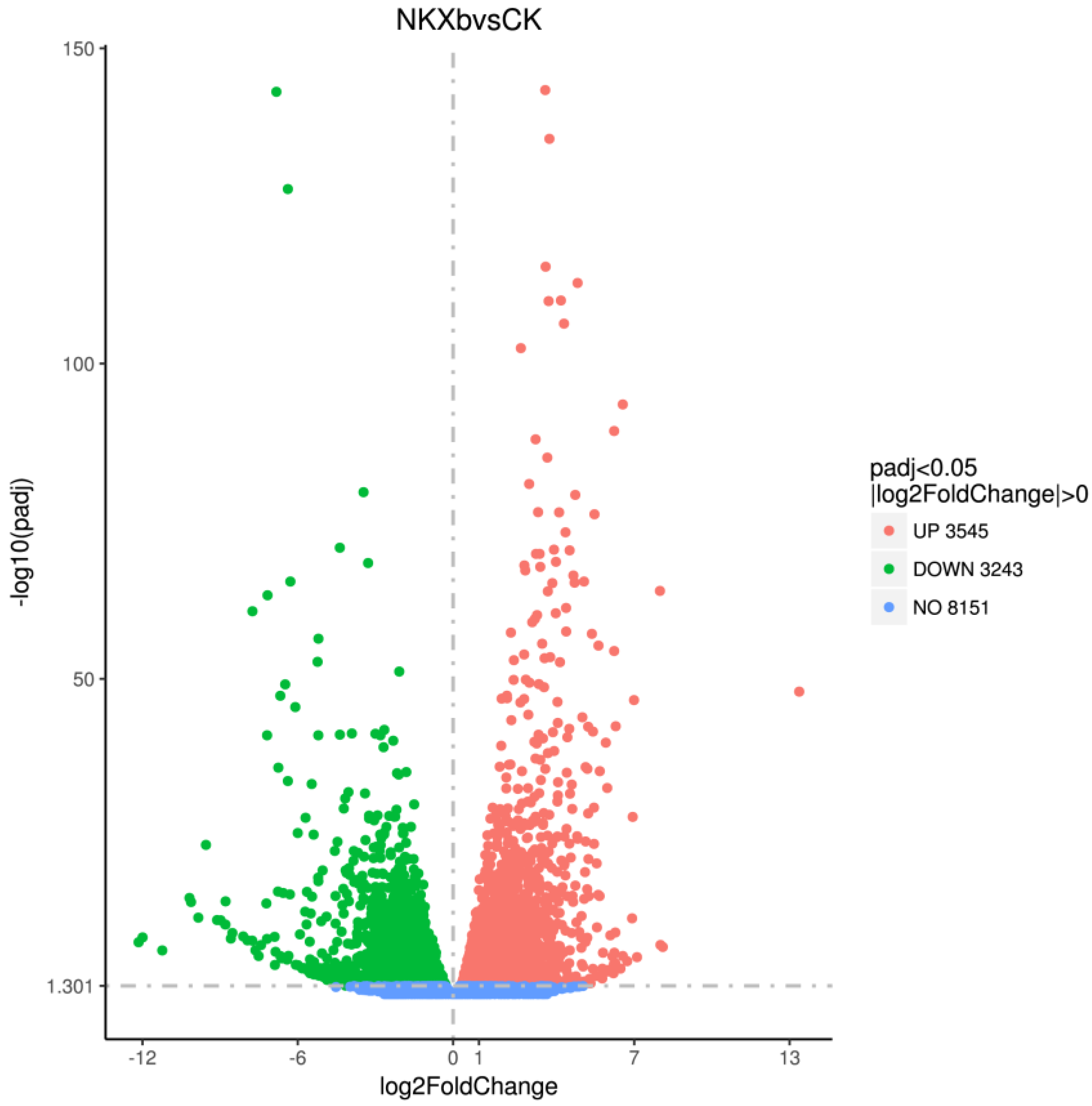
2.3. Transcriptomic Analysis of NK-NH1
2.3.1. Differential Expression Analysis
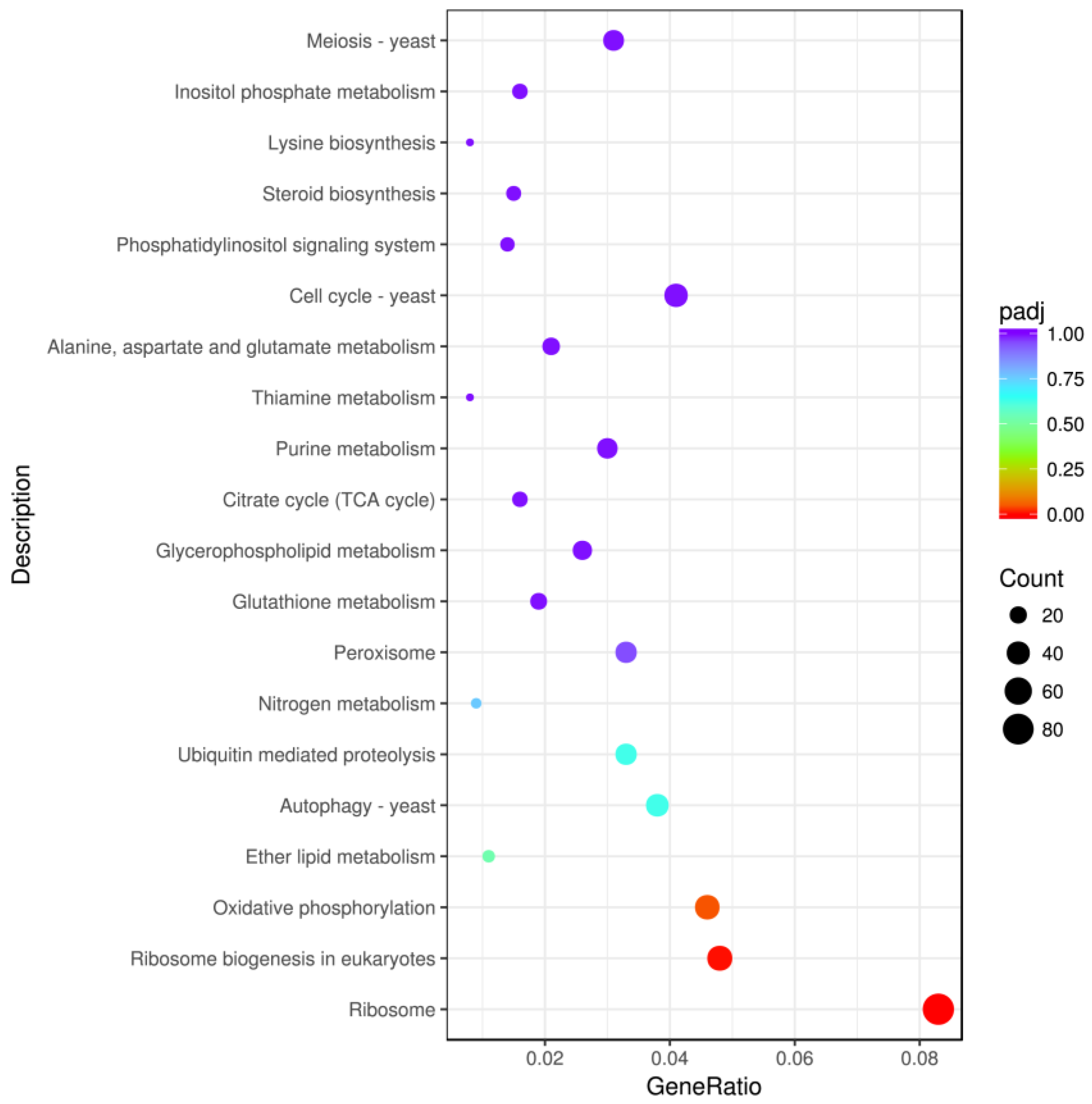
2.3.2. KEGG Enrichment Analysis of Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Strains
4.2. Inhibitory Effect of the Supernatant of X. bovienii Ferment on F. solani
4.3. Metabolome Analysis
4.3.1. Sample Preparation
4.3.2. Chromatography Conditions
4.3.3. Conditions Used for Mass Spectrometry
4.3.4. Data Analysis
4.4. Inhibitory Effect of VOCs Produced by X. bovienii on F. solani
4.5. Detection of VOCs Using GC-MS
4.5.1. Collection of VOCs Produced by X. bovienii
4.5.2. GC–MS Detection
4.6. Transcriptome Sequencing Analysis
4.6.1. Differential Expression Analysis
4.6.2. KEGG Enrichment Analysis of Differentially Expressed Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dillman, A.; Sternberg, P. Entomopathogenic nematodes. Curr. Biol. 2012, 22, 430–431. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Wouts, W.; Li, L. Development and virulence of Heterorhabditis spp. strains associated with different Xenorhabdus luminescens isolates. J. Invertebr. Pathol. 1991, 58, 27–32. [Google Scholar] [CrossRef]
- Kergunteuil, A.; Campos-Herrera, R.; Sánchez-Moreno, S.; Vittoz, P.; Rasmann, S. The abundance, diversity, and metabolic footprint of soil nematodes is highest in high elevation alpine grasslands. Front. Ecol. Evol. 2016, 4, 84–87. [Google Scholar] [CrossRef]
- Maher, A.; Asaiyah, M.; Brophy, C.; Griffin, C.T. An entomopathogenic nematode extends its niche by associating with different symbionts. Microb. Ecol. 2017, 73, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, R.-U. Current and future use of nematodes in biocontrol: Practice and commercial aspects with regard to regulatory policy issues. Biocontrol Sci. Technol. 1996, 6, 303–316. [Google Scholar] [CrossRef]
- Ehlers, R.-U.; Hokkanen, H.M.T. Insect biocontrol with non-endemic entomopathogenic nematodes (Steinernema and Heterorhabditis spp.): Conclusions and recommendations of a combined OECD and COST Workshop on Scientific and Regulatory Policy Issues. Biocontrol Sci. Technol. 1996, 6, 295–302. [Google Scholar] [CrossRef]
- Fukruksa, C.; Yimthin, T.; Suwannaroj, M.; Muangpat, P.; Tandhavanant, S.; Thanwisai, A. Isolation and identification of Xenorhabdus and Photorhabdus bacteria associated with entomopathogenic nematodes and their larvicidal activity against Aedes aegypti. Parasites Vectors 2017, 10, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Bozhüyük, K.; Zhou, Q.; Engel, Y.; Heinrich, A.; Pérez, A.; Bode, H. Natural Products from Photorhabdus and Other Entomopathogenic Bacteria. Curr. Top. Microbiol. Immunol. 2016, 24, 55–79. [Google Scholar]
- Maxwell, P.; Chen, G.; Webster, J.; Dunyhy, G. Culture conditions for Xenorhabdus and Photorhabdus symbionts of entomopathogenic nematodes. Nematologica 1996, 42, 124–130. [Google Scholar]
- Qiu, X.; Han, R.; Yan, X.; Liu, M.; Cao, L.; Yoshiga, T. Identification and characterization of a novel gene involved in the trans-specific nematicidal activity of Photorhabdus luminescens LN2. Appl. Environ. Microbiol. 2009, 75, 4221–4223. [Google Scholar] [CrossRef] [PubMed]
- McInerney, B.; Taylor, W.C.; Lacey, M.J.; Akhurst, R.J.; Gregson, R.P. Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 1991, 54, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Shapiro-Ilan, D.I.; Gouge, D.; Piggott, S.; Fife, J. Application technology and environmental considerations for use of entomopathogenic nematodes in biological control. Biol. Control 2006, 38, 124–133. [Google Scholar] [CrossRef]
- Ng, K.; Webster, J. Antimycotic activity of Xenorhabdus bovienii (Enterobacteriaceae) metabolites against Phytophthora infestans on potato plants. Can. J. Plant Pathol. 1997, 19, 125–132. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, X.; An, F.; Wang, G.; Zhang, X. Improvement of antibiotic activity of Xenorhabdus bovienii by medium optimization using response surface methodology. Microb. Cell Fact. 2011, 10, 98. [Google Scholar] [CrossRef]
- Paul, V.; Frautschy, S.; Fenical, W.; Nealson, K. Antibiotics in microbial ecology: Isolation and structure assignment of several new antibacterial compounds from the insect-symbiotic bacteria Xenorhabdus spp. J. Chem. Ecol. 1981, 7, 589–597. [Google Scholar] [CrossRef]
- McInerney, B.V.; Gregson, R.P.; Lacey, M.J.; Akhurst, R.J.; Lyons, G.R.; Rhodes, S.H.; Smith, D.R.J.; Engelhardt, L.M.; White, A.H. Biologically active metabolites from Xenorhabdus Spp., Part 1. Dithiolopyrrolone derivatives with antibiotic activity. J. Nat. Prod. 1991, 54, 774–784. [Google Scholar] [CrossRef]
- Ji, D.; Yi, Y.; Kang, G.; Choi, Y.; Kim, P.; Baek, N. Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol. Lett. 2006, 239, 241–248. [Google Scholar] [CrossRef]
- Fernando, W.; Ramarathnam, R.; Krishnamoorthy, A.; Savchuk, S. Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 2005, 37, 955–964. [Google Scholar] [CrossRef]
- Cappitelli, F.; Pasquariello, G.; Tarsitani, G. Assessing microbial risk to paper heritage. Trends Microbiol. 2010, 18, 538–542. [Google Scholar] [CrossRef]
- Sterflinger, K.; Piñar, G. Microbial deterioration of cultural heritage and works of art—Tilting at windmills. Appl. Microbiol. Biotechnol. 2013, 97, 9637–9646. [Google Scholar] [CrossRef]
- Jurado, V.; Miller, A.; Cuezva, S.; Fernandez-Cortes, A.; Benavente, D.; Rogerio-Candelera, M. Recolonization of mortars by endolithic organisms on the walls of San Roque church in Campeche (Mexico): A case of tertiary bioreceptivity. Constr. Build. Mater. 2014, 53, 348–359. [Google Scholar] [CrossRef]
- Diaz-Herraiz, M.; Jurado, V.; Cuezva, S.; Laiz, L.; Pallecchi, P.; Tiano, P. The actinobacterial colonization of Etruscan paintings. Sci. Rep. 2013, 3, 1440. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R.; Giuffrida, M.; Pessione, E. Back to the past: “find the guilty bug-microorganisms involved in the biodeterioration of archeological and historical artifacts”. Appl. Microbiol. Biotechnol. 2018, 102, 6393–6407. [Google Scholar] [CrossRef] [PubMed]
- Gutarowska, B.; Pietrzak, K.; Machnowski, W.; Miczarek, M. Historical textiles—A review of microbial deterioration analysis and disinfection methods. Text. Res. J. 2017, 87, 2388–2406. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Zhang, F. Microbial community analyses of the deteriorated storeroom objects in the Tianjin Museum using culture-independent and culture-dependent approaches. Front. Microbiol. 2018, 9, 802. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; Yang, B. Deterioration-associated microbiome of stone monuments: Structure, variation, and assembly. Appl. Environ. Microbiol. 2018, 84, e02680-17. [Google Scholar] [CrossRef]
- Han, Y.; Huang, X.; Wang, Y.; Du, J.; Ma, K.; Chen, Y.; Li, N.; Zhang, Z.; Pan, J. Fungal community and biodeterioration analysis of hull wood and its storage environment of the Nanhai No. 1 Shipwreck. Front. Microbiol. 2021, 11, 609475. [Google Scholar] [CrossRef]
- Houard, J.; Aumelas, A.; Noël, T.; Pages, S.; Gualtieri, M. Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J. Antibiot. 2013, 66, 617–620. [Google Scholar] [CrossRef]
- Bailly, A.; Weisskopf, L. Mining the volatilomes of plant-associated microbiota for new biocontrol solutions. Front. Microbiol. 2017, 8, 1638. [Google Scholar] [CrossRef]
- Blom, D.; Fabbri, C.; Eberl, L.; Weisskopf, L. Volatile-mediated killing of Arabidopsis thaliana by bacteria is mainly mediated due to hydrogen cyanide. Appl. Environ. Microbiol. 2011, 77, 1000–1008. [Google Scholar] [CrossRef]
- Schenkel, D.; Lemfack, M.C.; Piechulla, B.; Splivallo, R. A meta-analysis approach for assessing the diversity and specificity of belowground root and microbial volatiles. Front. Plant Sci. 2015, 6, 707. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Wang, W.; Song, C.; Wang, M.; Sun, B.; Li, Y. The symbiotic bacteria Alcaligenes faecalis of the entomopathogenic nematodes Oscheius spp. exhibit potential biocontrol of plant- and entomopathogenic fungi. Microb. Biotechnol. 2019, 12, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Zaman, S.; Broach, J.; Klionsky, D. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 4180–4189. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.; Bonenfant, D. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Crespo, J.L.; Powers, T.; Fowler, B.; Hall, M.N. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 2002, 99, 6784–6789. [Google Scholar] [CrossRef]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signaling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef]
- Budovskaya, Y.V.; Stephan, J.S.; Reggiori, F.; Klionsky, D.J.; Herman, P.K. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 20663–20671. [Google Scholar] [CrossRef]
- Budovskaya, Y.V.; Stephan, J.S.; Deminoff, S.J.; Herman, P.K. An evolutionary proteomics approach identifies substrates of the c AMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 2005, 102, 13933–13938. [Google Scholar] [CrossRef]
| Metabolite | Formula | FC (NKSF/Blank) | p Value |
|---|---|---|---|
| lysoPC (28:0) | C36H74NO7P | 4643.192488 | 1.47 × 10−14 |
| N-Carbamoylputrescine | C5H13N3O | 565,313.1885 | 2.67 × 10−11 |
| 20-HETE ethanolamide | C22H37NO3 | 462.7804571 | 9.11 × 10−11 |
| PE-NMe (16:0/22:5(4Z,7Z,10Z,13Z,16Z)) | C44H78NO8P | 403.5201482 | 3.26 × 10−14 |
| alpha-Ionol O-[arabinosyl-(1->6)-glucoside] | C24H40O10 | 525.0169799 | 4.03 × 10−8 |
| PG (16:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | C44H75O10P | 269.3971166 | 6.03 × 10−10 |
| PA (16:0/18:1(11Z)) | C37H71O8P | 108.0167793 | 3.29 × 10−9 |
| PA (16:0/15:0) | C34H67O8P | 105.6211958 | 1.09 × 10−5 |
| Lucidenic acid M | C27H42O6 | 25,115.625 | 1.36 × 10−6 |
| Validamycin A | C20H35NO13 | 1875.077176 | 1.74 × 10−7 |
| Octadecyl fumarate | C22H40O4 | 263.6363636 | 3.50 × 10−15 |
| Lucyoside R | C36H58O11 | 196.4816356 | 7.89 × 10−12 |
| 2,6,6-Trimethyl-1,4-cyclohexadiene-1-carboxaldehyde | C10H14O | 186.2414338 | 1.39 × 10−9 |
| 1-(2,3-Dihydro-1H-pyrrolizin-5-yl)-2-propen-1-one | C10H11NO | 6173.232908 | 4.04 × 10−10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, F.; Wang, C.; Guo, P.; Han, Y.; Zhang, Y.; Sun, B.; Shan, S.; Ruan, W.; Pan, J. Antifungal Substances Produced by Xenorhabdus bovienii and Its Inhibition Mechanism against Fusarium solani. Int. J. Mol. Sci. 2022, 23, 9040. https://doi.org/10.3390/ijms23169040
Wang Y, Zhang F, Wang C, Guo P, Han Y, Zhang Y, Sun B, Shan S, Ruan W, Pan J. Antifungal Substances Produced by Xenorhabdus bovienii and Its Inhibition Mechanism against Fusarium solani. International Journal of Molecular Sciences. 2022; 23(16):9040. https://doi.org/10.3390/ijms23169040
Chicago/Turabian StyleWang, Yu, Fengyu Zhang, Cen Wang, Peifeng Guo, Yeqing Han, Yingting Zhang, Bingjiao Sun, Shaojie Shan, Weibin Ruan, and Jiao Pan. 2022. "Antifungal Substances Produced by Xenorhabdus bovienii and Its Inhibition Mechanism against Fusarium solani" International Journal of Molecular Sciences 23, no. 16: 9040. https://doi.org/10.3390/ijms23169040
APA StyleWang, Y., Zhang, F., Wang, C., Guo, P., Han, Y., Zhang, Y., Sun, B., Shan, S., Ruan, W., & Pan, J. (2022). Antifungal Substances Produced by Xenorhabdus bovienii and Its Inhibition Mechanism against Fusarium solani. International Journal of Molecular Sciences, 23(16), 9040. https://doi.org/10.3390/ijms23169040





