A Metabolomic and Transcriptomic Study Revealed the Mechanisms of Lumefantrine Inhibition of Toxoplasma gondii
Abstract
1. Introduction
2. Results
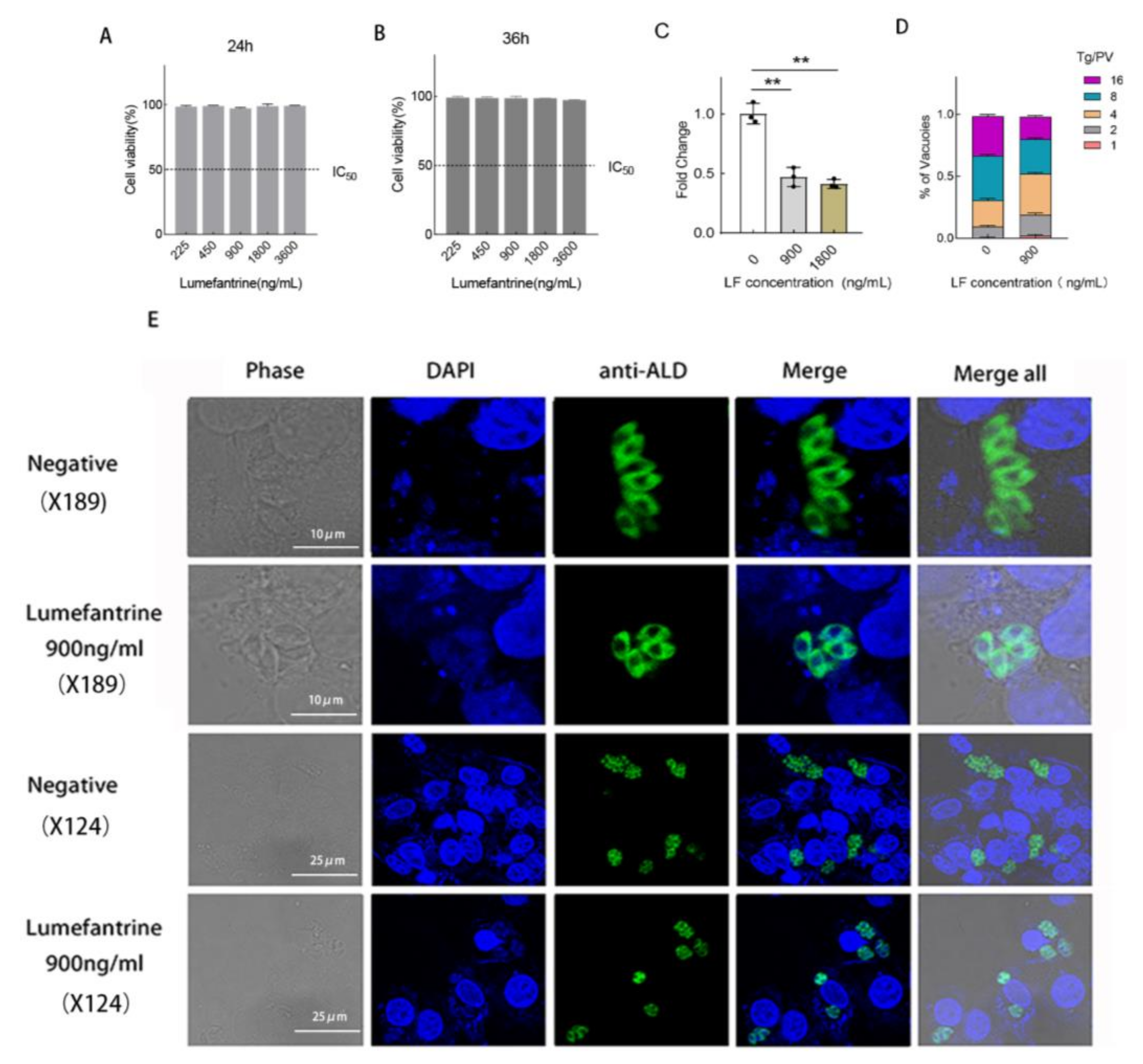
2.1. Lumefantrine Restricts the Proliferation of T. gondii
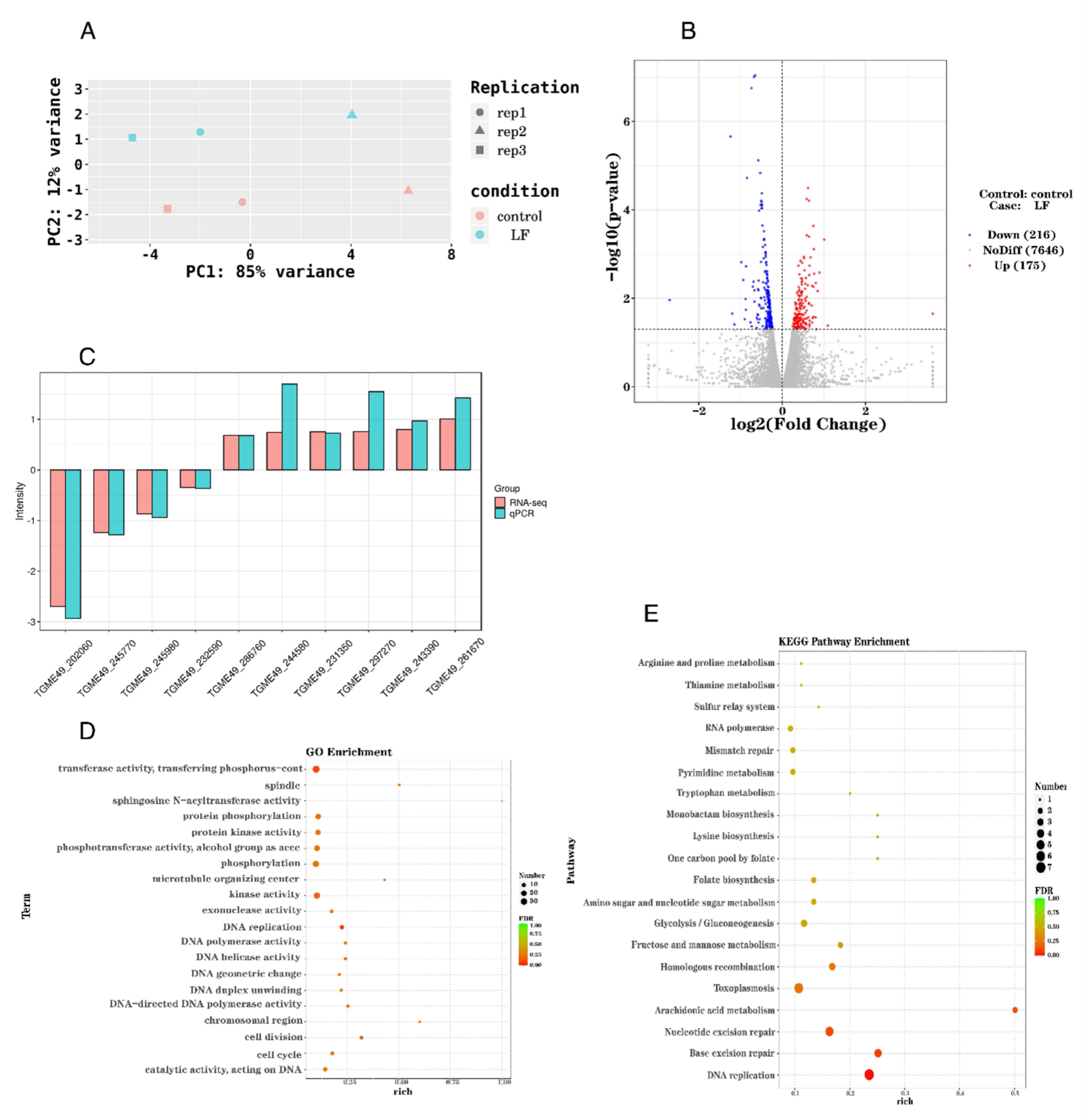
2.2. Lumefantrine Alters T. gondii Gene Expression
2.3. The Upregulation of Seven Genes Involved in DNA Replication
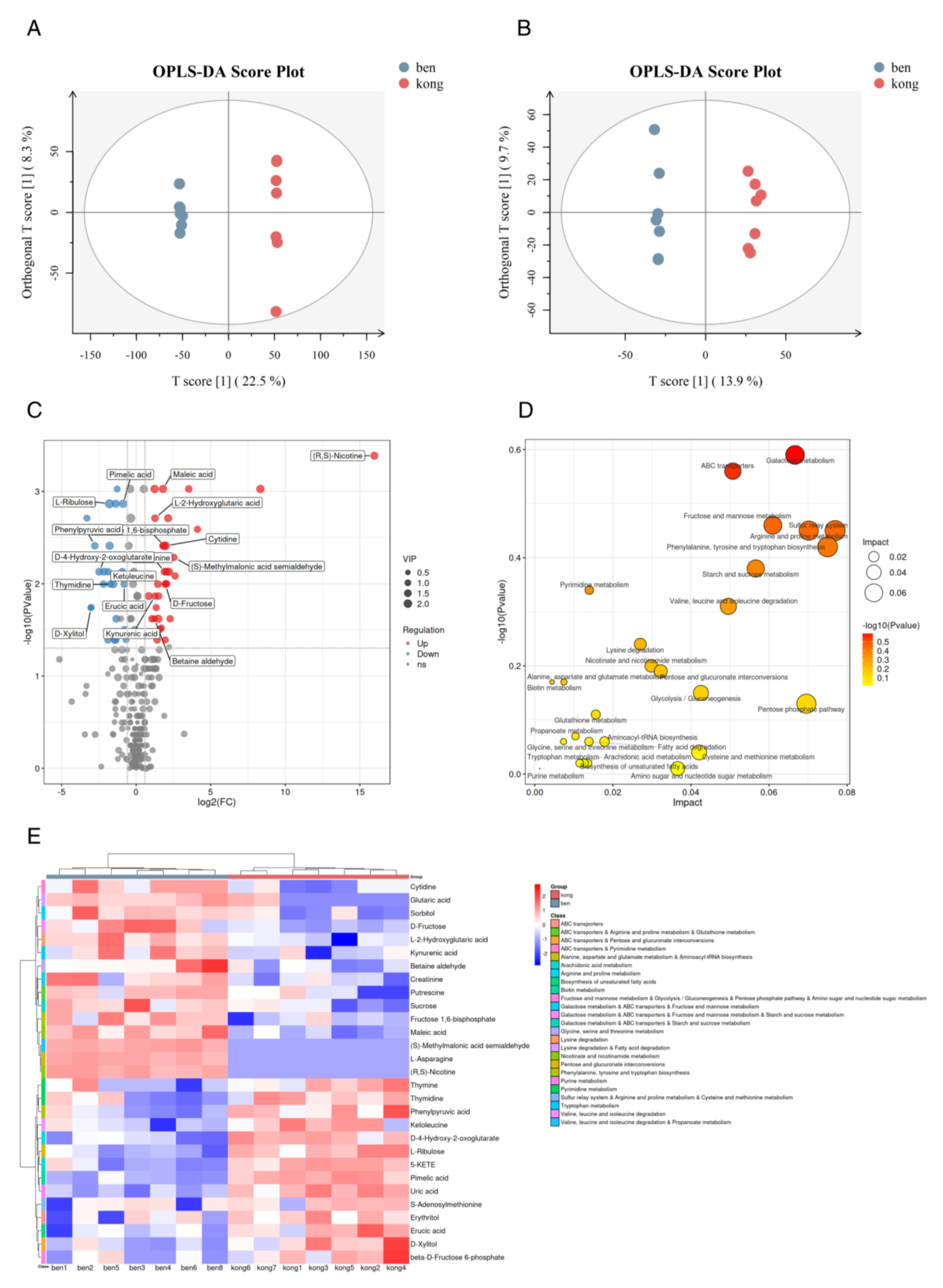
2.4. Lumefantrine Interfered with T. gondii Metabolism
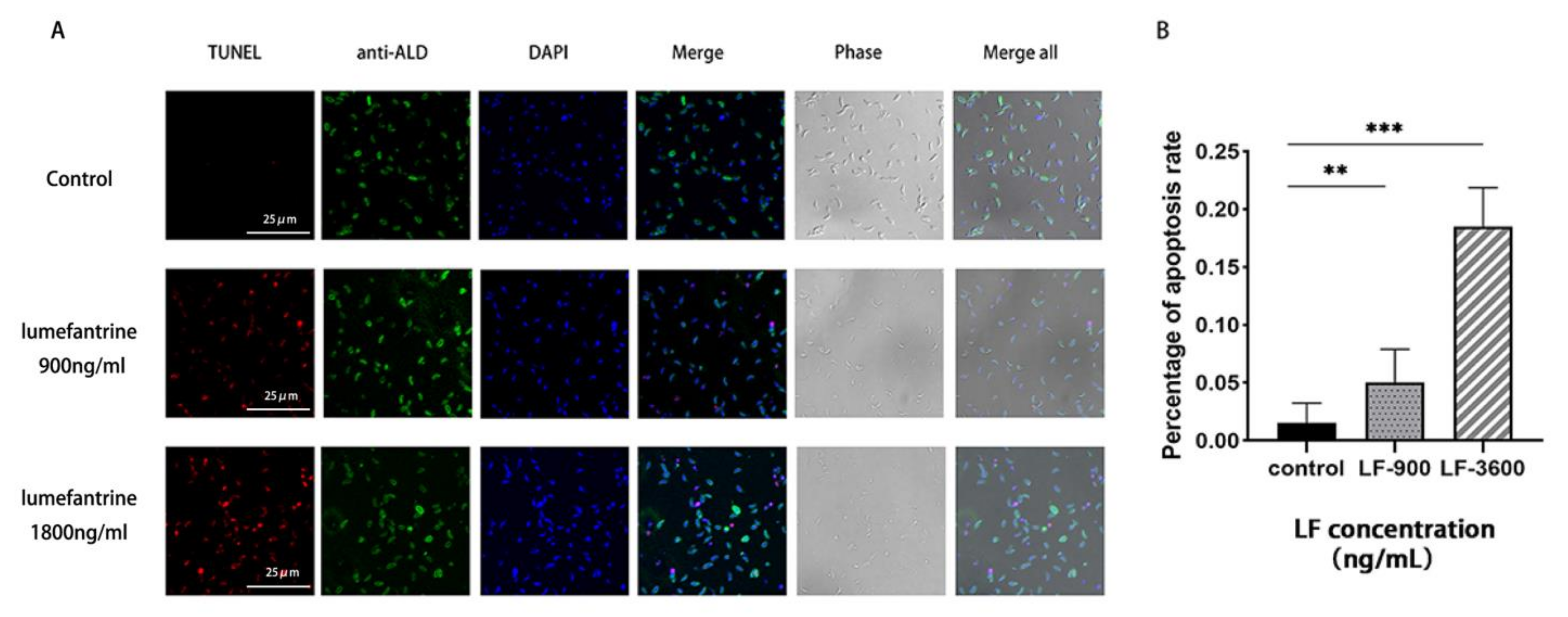
2.5. Lumefantrine-Induced T. gondii Apoptosis
3. Discussion
4. Materials and Methods
4.1. Cells and Parasites
4.2. Cytotoxicity Assay
4.3. Intracellular Proliferation Assay
4.4. In Vitro Proliferation Assay
4.5. Transcriptome Sample Preparation
4.6. Transcriptome Data Validation
4.7. Metabolome Sample Preparation and Metabolite Extraction
4.8. LC-MS/MS Analysis
4.9. Lumefantrine-Induced DNA Damage
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molan, A.; Nosaka, K.; Hunter, M.; Wang, W. Global status of Toxoplasma gondii infection: Systematic review and prevalence snapshots. Trop Biomed. 2019, 36, 898–925. [Google Scholar] [PubMed]
- Zhou, P.; Chen, Z.; Li, H.L.; Zheng, H.; He, S.; Lin, R.Q.; Zhu, X.Q. Toxoplasma gondii infection in humans in China. Parasit Vectors 2011, 4, 165. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, H.W.; Huang, K.Q.; Xu, Y.H.; Li, Y.N.; Du, J.; Yu, L.; Luo, Q.L.; Wei, W.; Jiang, L.; et al. Toxoplasma gondii prevalence in food animals and rodents in different regions of China: Isolation, genotyping and mouse pathogenicity. Parasit Vectors 2013, 6, 273. [Google Scholar] [CrossRef]
- Zhou, D.H.; Zhao, F.R.; Huang, S.Y.; Xu, M.J.; Song, H.Q.; Su, C.; Zhu, X.Q. Changes in the proteomic profiles of mouse brain after infection with cyst-forming Toxoplasma gondii. Parasit Vectors 2013, 6, 96. [Google Scholar] [CrossRef]
- Qin, S.Y.; Cong, W.; Liu, Y.; Li, N.; Wang, Z.D.; Zhang, F.K.; Huang, S.Y.; Zhu, X.Q.; Liu, Q. Molecular detection and genotypic characterization of Toxoplasma gondii infection in bats in four provinces of China. Parasit Vectors 2014, 7, 558. [Google Scholar] [CrossRef]
- Wang, L.; He, L.Y.; Meng, D.D.; Chen, Z.W.; Wen, H.; Fang, G.S.; Luo, Q.L.; Huang, K.Q.; Shen, J.L. Seroprevalence and genetic characterization of Toxoplasma gondii in cancer patients in Anhui Province, Eastern China. Parasit Vectors 2015, 8, 162. [Google Scholar] [CrossRef]
- Rorman, E.; Zamir, C.S.; Rilkis, I.; Ben-David, H. Congenital toxoplasmosis--prenatal aspects of Toxoplasma gondii infection. Reprod. Toxicol. 2006, 21, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Fallahi, S.; Rostami, A.; Nourollahpour Shiadeh, M.; Behniafar, H.; Paktinat, S. An updated literature review on maternal-fetal and reproductive disorders of Toxoplasma gondii infection. J. Gynecol. Obstet. Hum. Reprod 2018, 47, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Flegr, J. How and why Toxoplasma makes us crazy. Trends Parasitol. 2013, 29, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Fond, G.; Capdevielle, D.; Macgregor, A.; Attal, J.; Larue, A.; Brittner, M.; Ducasse, D.; Boulenger, J.P. Toxoplasma gondii: A potential role in the genesis of psychiatric disorders. L’Encephale 2013, 39, 38–43. [Google Scholar] [CrossRef]
- Berger-Schoch, A.E.; Herrmann, D.C.; Schares, G.; Müller, N.; Bernet, D.; Gottstein, B.; Frey, C.F. Prevalence and genotypes of Toxoplasma gondii in feline faeces (oocysts) and meat from sheep, cattle and pigs in Switzerland. Vet. Parasitol. 2011, 177, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.A.; Longcore, T.; Barbieri, M.; Dabritz, H.; Hill, D.; Klein, P.N.; Lepczyk, C.; Lilly, E.L.; McLeod, R.; Milcarsky, J.; et al. The One Health Approach to Toxoplasmosis: Epidemiology, Control, and Prevention Strategies. Ecohealth 2019, 16, 378–390. [Google Scholar] [CrossRef] [PubMed]
- Araujo, F.G.; Suzuki, Y.; Remington, J.S. Use of rifabutin in combination with atovaquone, clindamycin, pyrimethamine, or sulfadiazine for treatment of toxoplasmic encephalitis in mice. Eur. J. Clin. Microbiol. Infect. Dis. 1996, 15, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.G.; Liesenfeld, O. Toxoplasmosis. Lancet 2004, 363, 1965–1976. [Google Scholar] [CrossRef]
- Alves, C.F.; Vitor, R.W. Efficacy of atovaquone and sulfadiazine in the treatment of mice infected with Toxoplasma gondii strains isolated in Brazil. Parasite 2005, 12, 171–177. [Google Scholar] [CrossRef]
- Meneceur, P.; Bouldouyre, M.A.; Aubert, D.; Villena, I.; Menotti, J.; Sauvage, V.; Garin, J.F.; Derouin, F. In vitro susceptibility of various genotypic strains of Toxoplasma gondii to pyrimethamine, sulfadiazine, and atovaquone. Antimicrob. Agents Chemother. 2008, 52, 1269–1277. [Google Scholar] [CrossRef]
- Rajapakse, S.; Chrishan Shivanthan, M.; Samaranayake, N.; Rodrigo, C.; Deepika Fernando, S. Antibiotics for human toxoplasmosis: A systematic review of randomized trials. Pathog. Glob. Health 2013, 107, 162–169. [Google Scholar] [CrossRef]
- Neville, A.J.; Zach, S.J.; Wang, X.; Larson, J.J.; Judge, A.K.; Davis, L.A.; Vennerstrom, J.L.; Davis, P.H. Clinically Available Medicines Demonstrating Anti-Toxoplasma Activity. Antimicrob. Agents Chemother. 2015, 59, 7161–7169. [Google Scholar] [CrossRef]
- Deng, Y.; Wu, T.; Zhai, S.Q.; Li, C.H. Recent progress on anti-Toxoplasma drugs discovery: Design, synthesis and screening. Eur. J. Med. Chem. 2019, 183, 111711. [Google Scholar] [CrossRef]
- Shiojiri, D.; Kinai, E.; Teruya, K.; Kikuchi, Y.; Oka, S. Combination of Clindamycin and Azithromycin as Alternative Treatment for Toxoplasma gondii Encephalitis. Emerg. Infect Dis. 2019, 25, 841–843. [Google Scholar] [CrossRef]
- Shah, R.; Soni, T.; Shah, U.; Suhagia, B.N.; Patel, M.N.; Patel, T.; Gabr, G.A.; Gorain, B.; Kesharwani, P. Formulation development and characterization of lumefantrine nanosuspension for enhanced antimalarial activity. J. Biomat. Sci. Polym. E 2021, 32, 833–857. [Google Scholar] [CrossRef] [PubMed]
- Wernsdorfer, W.H. Coartemether (artemether and lumefantrine): An oral antimalarial drug. Expert Rev. Anti Infect Ther. 2004, 2, 181–196. [Google Scholar] [CrossRef] [PubMed]
- Stover, K.R.; King, S.T.; Robinson, J. Artemether-lumefantrine: An option for malaria. Ann. Pharm. 2012, 46, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xing, M.; El-Ashram, S.; Ding, Y.; Zhang, X.; Sang, X.; Feng, Y.; Chen, R.; Wang, X.; Jiang, N.; et al. Determination of lumefantrine as an effective drug against Toxoplasma gondii infection—In vitro and in vivo study. Parasitology 2021, 148, 122–128. [Google Scholar] [CrossRef]
- Costa, V.; Angelini, C.; De Feis, I.; Ciccodicola, A. Uncovering the complexity of transcriptomes with RNA-Seq. J. Biomed. Biotechnol. 2010, 2010, 853916. [Google Scholar] [CrossRef]
- Finotello, F.; Di Camillo, B. Measuring differential gene expression with RNA-seq: Challenges and strategies for data analysis. Brief Funct. Genom. 2015, 14, 130–142. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Zeki, Ö.C.; Eylem, C.C.; Reçber, T.; Kır, S.; Nemutlu, E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020, 190, 113509. [Google Scholar] [CrossRef]
- Tewari, S.G.; Kwan, B.; Elahi, R.; Rajaram, K.; Reifman, J.; Prigge, S.T.; Vaidya, A.B.; Wallqvist, A. Metabolic adjustments of blood-stage Plasmodium falciparum in response to sublethal pyrazoleamide exposure. Sci. Rep. 2022, 12, 1167. [Google Scholar] [CrossRef]
- Jia, Z.; Zhang, Z.; Tian, Q.; Wu, H.; Xie, Y.; Li, A.; Zhang, H.; Yang, Z.; Zhang, X. Integration of transcriptomics and metabolomics reveals anlotinib-induced cytotoxicity in colon cancer cells. Gene 2021, 786, 145625. [Google Scholar] [CrossRef]
- Dewar, J.M.; Walter, J.C. Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 2017, 18, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.B.; Klungland, A.; Rognes, T.; Leiros, I. DNA repair in mammalian cells: Base excision repair: The long and short of it. Cell Mol. Life Sci. 2009, 66, 981–993. [Google Scholar] [CrossRef] [PubMed]
- van Hoffen, A.; Balajee, A.S.; van Zeeland, A.A.; Mullenders, L.H. Nucleotide excision repair and its interplay with transcription. Toxicology 2003, 193, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Apostolou, Z.; Chatzinikolaou, G.; Stratigi, K.; Garinis, G.A. Nucleotide Excision Repair and Transcription-Associated Genome Instability. Bioessays 2019, 41, e1800201. [Google Scholar] [CrossRef] [PubMed]
- Lima, T.S.; Mallya, S.; Jankeel, A.; Messaoudi, I.; Lodoen, M.B. Toxoplasma gondii Extends the Life Span of Infected Human Neutrophils by Inducing Cytosolic PCNA and Blocking Activation of Apoptotic Caspases. mBio 2021, 12, e02031-20. [Google Scholar] [CrossRef] [PubMed]
- Forsburg, S.L. Eukaryotic MCM proteins: Beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004, 68, 109–131. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, Y.; Yang, X.; Liu, J.; Liu, Q. Toxoplasma gondii UBL-UBA shuttle proteins regulate several important cellular processes. FASEB J. 2021, 35, e21898. [Google Scholar] [CrossRef]
- Prosperi, E. Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair and cell cycle control. Prog. Cell Cy Res. 1997, 3, 193–210. [Google Scholar]
- Gupta, D.K.; Patra, A.T.; Zhu, L.; Gupta, A.P.; Bozdech, Z. DNA damage regulation and its role in drug-related phenotypes in the malaria parasites. Sci. Rep. 2016, 6, 23603. [Google Scholar] [CrossRef]
- Xiong, A.; Prakash, P.; Gao, X.H.; Chew, M.; Tay, I.J.J.; Woodrow, C.J.; Engelward, B.P.; Han, J.Y.; Preiser, P.R. K13-Mediated Reduced Susceptibility to Artemisinin in Plasmodium falciparum Is Overlaid on a Trait of Enhanced DNA Damage Repair. Cell Rep. 2020, 5, 107996. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, A.M.; Kumar, N. Antimalarial Action of Artesunate Involves DNA Damage Mediated by Reactive Oxygen Species. Antimicrob. Agents Chem. 2015, 1, 317–325. [Google Scholar] [CrossRef]
- Rodrigo, M.E.; Raúl, A.G.; Emma, S.; Guadalupe, O.P. Albendazole induces oxidative stress and DNA damage in the parasitic protozoan Giardia duodenalis. Front. Microbiol. 2015, 6, 800. [Google Scholar]
- Maréchal, E.; Azzouz, N.; de Macedo, C.S.; Block, M.A.; Feagin, J.E.; Schwarz, R.T.; Joyard, J. Synthesis of chloroplast galactolipids in apicomplexan parasites. Eukaryotic Cell 2002, 1, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Kavishe, R.A.; van den Heuvel, J.M.; van de Vegte-Bolmer, M.; Luty, A.J.; Russel, F.G.; Koenderink, J.B. Localization of the ATP-binding cassette (ABC) transport proteins PfMRP1, PfMRP2, and PfMDR5 at the Plasmodium falciparum plasma membrane. Malar. J. 2009, 8, 205. [Google Scholar] [CrossRef]
- Valderramos, S.G.; Fidock, D.A. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 2006, 27, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Demidenko, R.; Razanauskas, D.; Daniunaite, K.; Lazutka, J.R.; Jankevicius, F.; Jarmalaite, S. Frequent down-regulation of ABC transporter genes in prostate cancer. BMC Cancer 2015, 15, 683. [Google Scholar] [CrossRef]
- Larrazabal, C.; Silva, L.; Pervizaj-Oruqaj, L.; Herold, S.; Hermosilla, C.; Taubert, A. P-Glycoprotein Inhibitors Differently Affect Toxoplasma gondii, Neospora caninum and Besnoitia besnoiti Proliferation in Bovine Primary Endothelial Cells. Pathogens 2021, 10, 395. [Google Scholar] [CrossRef]
- Blume, M.; Rodriguez-Contreras, D.; Landfear, S.; Fleige, T.; Soldati-Favre, D.; Lucius, R.; Gupta, N. Host-derived glucose and its transporter in the obligate intracellular pathogen Toxoplasma gondii are dispensable by glutaminolysis. Proc. Natl. Acad. Sci. USA 2009, 106, 12998–13003. [Google Scholar] [CrossRef]
- Krishnan, A.; Soldati-Favre, D. Amino Acid Metabolism in Apicomplexan Parasites. Metabolites 2021, 11, 61. [Google Scholar] [CrossRef]
- Hargrave, K.E.; Woods, S.; Millington, O.; Chalmers, S.; Westrop, G.D.; Roberts, C.W. Multi-Omics Studies Demonstrate Toxoplasma gondii-Induced Metabolic Reprogramming of Murine Dendritic Cells. Front. Cell Infect. Microbiol. 2019, 9, 309. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene Name | Forward Primer | Reverse Primer |
|---|---|---|---|
| TGME49_202060 | hypothetical protein | CCTCACCAAGGAATGACACGA | TATGACGCTGGAGCGAGGAA |
| TGME49_245770 | hypothetical protein | GAACATGCGAATGCACGAGA | ATGCGTTCCCCCACTTTTTG |
| TGME49_245980 | hypothetical protein | CCACGTGTAATGCGTGAGTTG | TCACACTGGTACGACAAGCAA |
| TGME49_232590 | glutamate-cysteine ligase, catalytic subunit domain-containing protein | CCCCTGGTGACAGCTGATTT | TTGGTCGTGATAGGGCGATG |
| TGME49_286760 | hypothetical protein | AACGGGTTGAGTGTGCAAGA | GTCGTTTGAGGCTGCTGTTG |
| TGME49_244580 | L1P family of ribosomal protein | AGCAGCCAGAGTGTTTCAGT | CAGAGCGCGGGAAGAGTAAA |
| TGME49_231350 | glucosamine-fructose-6-phosphate aminotransferase | GATCGCACCCTACCAGAACC | TGAGCAGTTCGCCGTAGATG |
| TGME49_297270 | hypothetical protein | TGGGGATACCCCATTTTCGG | TGTCTCATACTCCCAGGCCA |
| TGME49_243390 | hypothetical protein | ATCGAGGCATCATCAACCCC | CGGGTAGGCAAGGGATCAAA |
| TGME49_261670 | ribonuclease H1/H2 small subunit protein | CCACTTTCTCACCATGCCCT | GAACCATCCGTTTCCCCTGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Sang, X.; Zhang, X.; Li, X.; Feng, Y.; Yang, N.; Jiang, T. A Metabolomic and Transcriptomic Study Revealed the Mechanisms of Lumefantrine Inhibition of Toxoplasma gondii. Int. J. Mol. Sci. 2023, 24, 4902. https://doi.org/10.3390/ijms24054902
Li M, Sang X, Zhang X, Li X, Feng Y, Yang N, Jiang T. A Metabolomic and Transcriptomic Study Revealed the Mechanisms of Lumefantrine Inhibition of Toxoplasma gondii. International Journal of Molecular Sciences. 2023; 24(5):4902. https://doi.org/10.3390/ijms24054902
Chicago/Turabian StyleLi, Meiqi, Xiaoyu Sang, Xiaohan Zhang, Xiang Li, Ying Feng, Na Yang, and Tiantian Jiang. 2023. "A Metabolomic and Transcriptomic Study Revealed the Mechanisms of Lumefantrine Inhibition of Toxoplasma gondii" International Journal of Molecular Sciences 24, no. 5: 4902. https://doi.org/10.3390/ijms24054902
APA StyleLi, M., Sang, X., Zhang, X., Li, X., Feng, Y., Yang, N., & Jiang, T. (2023). A Metabolomic and Transcriptomic Study Revealed the Mechanisms of Lumefantrine Inhibition of Toxoplasma gondii. International Journal of Molecular Sciences, 24(5), 4902. https://doi.org/10.3390/ijms24054902








