- Article
Loss of HuD Sensitizes Neuroblastoma Cells to Palmitate-Driven Stress-Induced Premature Senescence via PPARα Downregulation and FAO Impairment
- Seungyeon Ryu,
- Jiyoon Seo and
- Eun Kyung Lee
- + 4 authors
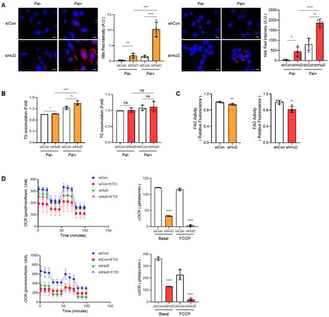
Metabolic stress caused by lipid overload is a key driver of cellular dysfunction in aging and disease. Excess saturated fatty acids such as palmitate impair fatty acid oxidation (FAO), promote lipid accumulation, and increase reactive oxygen species (ROS), ultimately triggering premature senescence-like states. Senescence further amplifies vulnerability by worsening mitochondrial dysfunction, enhancing lipid imbalance, and sustaining pro-inflammatory signaling. Here, we investigated the role of the neuron-enriched RNA-binding protein HuD (ELAVL4) in protecting cells against lipotoxic stress. Using Neuro2a neuroblastoma cells, we found that HuD knockdown suppressed FAO, leading to increased lipid accumulation and elevated ROS following palmitate exposure. HuD-deficient cells also exhibited cytosolic mitochondrial DNA release, IRF phosphorylation, and upregulation of senescence markers. Mechanistically, RNA immunoprecipitation revealed that HuD binds directly to PPARα mRNA, sustaining its expression by competing with the PPARα-targeting microRNAs miR-9-5p and miR-22-3p. Loss of HuD reduced PPARα levels, thereby weakening the FAO capacity and sensitizing cells to palmitate-induced lipotoxic stress. These findings identify a previously unrecognized HuD–PPARα–FAO axis that restrains metabolic stress and senescence. By linking post-transcriptional regulation to lipid metabolism and inflammatory signaling, this work highlights stress-induced premature senescence as both an outcome and a propagator of metabolic dysfunction, providing insight into mechanisms of aging-related vulnerability.
7 February 2026