- Article
PTMs_Closed_Search: Multiple Post-Translational Modification Closed Search Using Reduced Search Space and Transferred FDR
- Yury Yu. Strogov,
- Sergey A. Spirin and
- Oleg I. Klychnikov
- + 3 authors
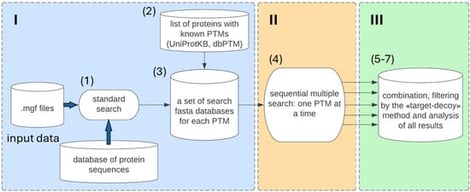
Background: Currently, post-translational modification (PTM) search in MS/MS data is performed using either open modification search (OMS) or closed search (CS) algorithms. The OMS method allows for the determination of many PTMs and unknown mass-shifts in one run. In contrast, closed search algorithms are more sensitive but limited in the number of PTMs that can be specified in one search. Methods: In this manuscript, we propose an optimized Python algorithm based on the IdentiPy search engine that performs an automated sequential search for each PTM based on previous annotations from public databases and customized protein lists. We also determined the sufficient size of the search space to increase the significance of false discovery rate (FDR) estimation. We modified the FDR calculation algorithm by implementing a spline approximation of the ratio of the modified decoys, and by calculating error propagation to filter out unstable data and determine the cutoff value. Results: The results of this pipeline for a test dataset were comparable to previously published data in terms of the number of unmodified peptides and proteins. Additionally, we identified 13 different types of peptide PTMs and achieved an increase in relative protein coverage. Our filtration method based on spline transferred FDR showed a superior number of identified peptides compared to separate FDR. Conclusions: Our developed pipeline can be used as a standalone application or as a module of multiple PTM search in data analysis platforms.
2 February 2026