Computational Imaging for Biophotonics and Biomedicine
A topical collection in Cells (ISSN 2073-4409). This collection belongs to the section "Cell Methods".
Viewed by 56505
Share This Topical Collection
Editors
 Prof. Dr. Baoli Yao
Prof. Dr. Baoli Yao
 Prof. Dr. Baoli Yao
Prof. Dr. Baoli Yao
E-Mail
Website
Collection Editor
State Key Laboratory of Transient Optics and Photonics, Xi'an Institute of Optics and Precision Mechanics, Chinese Academy Sciences, Xi’an 710119, China
Interests: super-resolution and 3D optical microscopy; quantitative phase imaging; optical trapping and manipulation
Special Issues, Collections and Topics in MDPI journals
 Prof. Dr. Chao Zuo
Prof. Dr. Chao Zuo
 Prof. Dr. Chao Zuo
Prof. Dr. Chao Zuo
grade
E-Mail
Website
Collection Editor
Department of Electronic and Optical Engineering, Nanjing University of Science and Technology, Nanjing 210094, China
Interests: computational bio-imaging; noninterferometic phase retrieval; optical information processing; high-speed 3D optical sensing
Special Issues, Collections and Topics in MDPI journals
 Dr. Jiamiao Yang
Dr. Jiamiao Yang
 Dr. Jiamiao Yang
Dr. Jiamiao Yang
E-Mail
Website
Collection Editor
Department of Instrument Science and Engineering, Shanghai Jiao Tong University, Shanghai 200240, China
Interests: photoacoustic imaging; wavefront shaping; scattering imaging and tomography
Topical Collection Information
Dear Colleagues,
Due to the improvement of advanced algorithms and computational power, computational imaging has experienced tremendous growth in recent years and has attracted the attention of many researchers, which may evolve conventional imaging technologies and bring many exciting opportunities for biophotonics and biomedicine. Therefore, we are writing to invite you to participate by submitting a manuscript to a Topical Collection of Cells (IF=6.6), entitled “Computational Imaging for Biophotonics and Biomedicine”. This Topical Collection plans to give an overview of the most recent advances in the field of computational imaging and their applications in biophotonics and biomedicine. The aim of this Topical Collection is to provide selected contributions on advances in both technology developments, as well as basic and clinical applications. Articles will be peer-reviewed and published in the open access journal Cells. Publication fee discounts are available for invitations. We look forward to your contributions.
Potential topics include, but are not limited to, the following:
- Advanced technologies in microscopy and spectroscopy;
- Quantitative phase imaging;
- Optical super-resolution imaging;
- Optofluidics and on-chip microscopy;
- Scattering imaging and tomography;
- Machine learning and AI.
Dr. An Pan
Prof. Dr. Baoli Yao
Prof. Dr. Chao Zuo
Dr. Fei Liu
Dr. Jiamiao Yang
Prof. Dr. Liangcai Cao
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cells is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- computational imaging
- biophotonics
- biomedicine
- microscopy
- spectroscopy
Published Papers (13 papers)
Open AccessEditorial
Computational Imaging: The Next Revolution for Biophotonics and Biomedicine
by
An Pan, Baoli Yao, Chao Zuo, Fei Liu, Jiamiao Yang and Liangcai Cao
Cited by 6 | Viewed by 2540
Abstract
This Editorial is the preface for the topical collection of “Computational Imaging for Biophotonics and Biomedicine”, which collates the 12 contributions listed in Table 1 [...]
Full article
Open AccessReview
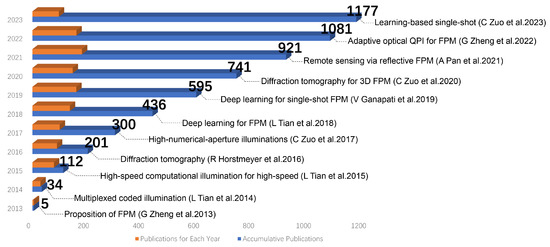
Fourier Ptychographic Microscopy 10 Years on: A Review
by
Fannuo Xu, Zipei Wu, Chao Tan, Yizheng Liao, Zhiping Wang, Keru Chen and An Pan
Cited by 24 | Viewed by 7782
Abstract
Fourier ptychographic microscopy (FPM) emerged as a prominent imaging technique in 2013, attracting significant interest due to its remarkable features such as precise phase retrieval, expansive field of view (FOV), and superior resolution. Over the past decade, FPM has become an essential tool
[...] Read more.
Fourier ptychographic microscopy (FPM) emerged as a prominent imaging technique in 2013, attracting significant interest due to its remarkable features such as precise phase retrieval, expansive field of view (FOV), and superior resolution. Over the past decade, FPM has become an essential tool in microscopy, with applications in metrology, scientific research, biomedicine, and inspection. This achievement arises from its ability to effectively address the persistent challenge of achieving a trade-off between FOV and resolution in imaging systems. It has a wide range of applications, including label-free imaging, drug screening, and digital pathology. In this comprehensive review, we present a concise overview of the fundamental principles of FPM and compare it with similar imaging techniques. In addition, we present a study on achieving colorization of restored photographs and enhancing the speed of FPM. Subsequently, we showcase several FPM applications utilizing the previously described technologies, with a specific focus on digital pathology, drug screening, and three-dimensional imaging. We thoroughly examine the benefits and challenges associated with integrating deep learning and FPM. To summarize, we express our own viewpoints on the technological progress of FPM and explore prospective avenues for its future developments.
Full article
►▼
Show Figures
Open AccessArticle
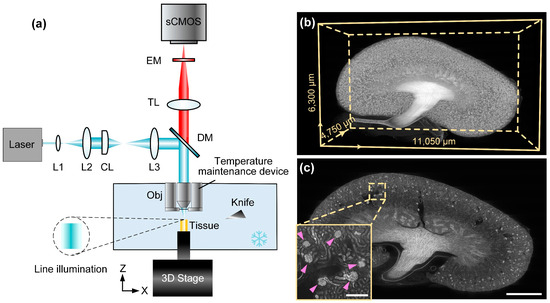
FastCellpose: A Fast and Accurate Deep-Learning Framework for Segmentation of All Glomeruli in Mouse Whole-Kidney Microscopic Optical Images
by
Yutong Han, Zhan Zhang, Yafeng Li, Guoqing Fan, Mengfei Liang, Zhijie Liu, Shuo Nie, Kefu Ning, Qingming Luo and Jing Yuan
Cited by 4 | Viewed by 2793
Abstract
Automated evaluation of all glomeruli throughout the whole kidney is essential for the comprehensive study of kidney function as well as understanding the mechanisms of kidney disease and development. The emerging large-volume microscopic optical imaging techniques allow for the acquisition of mouse whole-kidney
[...] Read more.
Automated evaluation of all glomeruli throughout the whole kidney is essential for the comprehensive study of kidney function as well as understanding the mechanisms of kidney disease and development. The emerging large-volume microscopic optical imaging techniques allow for the acquisition of mouse whole-kidney 3D datasets at a high resolution. However, fast and accurate analysis of massive imaging data remains a challenge. Here, we propose a deep learning-based segmentation method called FastCellpose to efficiently segment all glomeruli in whole mouse kidneys. Our framework is based on Cellpose, with comprehensive optimization in network architecture and the mask reconstruction process. By means of visual and quantitative analysis, we demonstrate that FastCellpose can achieve superior segmentation performance compared to other state-of-the-art cellular segmentation methods, and the processing speed was 12-fold higher than before. Based on this high-performance framework, we quantitatively analyzed the development changes of mouse glomeruli from birth to maturity, which is promising in terms of providing new insights for research on kidney development and function.
Full article
►▼
Show Figures
Open AccessArticle
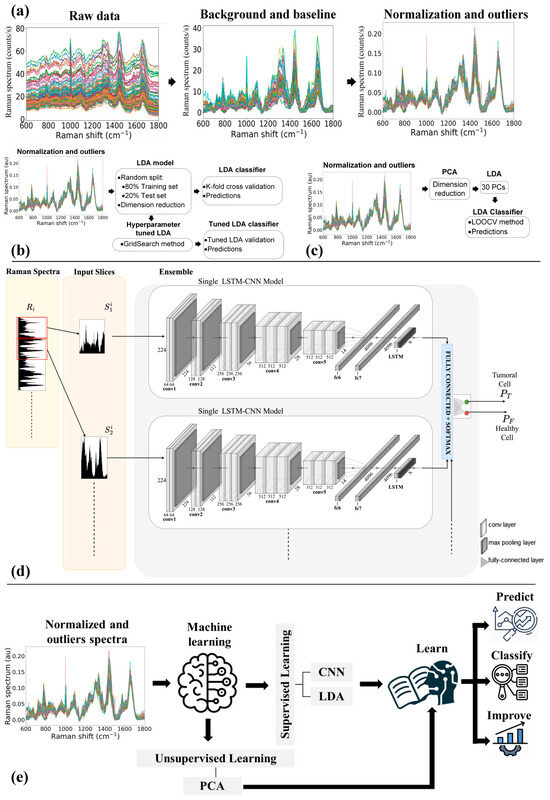
Assessment of Primary Human Liver Cancer Cells by Artificial Intelligence-Assisted Raman Spectroscopy
by
Concetta Esposito, Mohammed Janneh, Sara Spaziani, Vincenzo Calcagno, Mario Luca Bernardi, Martina Iammarino, Chiara Verdone, Maria Tagliamonte, Luigi Buonaguro, Marco Pisco, Lerina Aversano and Andrea Cusano
Cited by 13 | Viewed by 2831
Abstract
We investigated the possibility of using Raman spectroscopy assisted by artificial intelligence methods to identify liver cancer cells and distinguish them from their Non-Tumor counterpart. To this aim, primary liver cells (40 Tumor and 40 Non-Tumor cells) obtained from resected hepatocellular carcinoma (HCC)
[...] Read more.
We investigated the possibility of using Raman spectroscopy assisted by artificial intelligence methods to identify liver cancer cells and distinguish them from their Non-Tumor counterpart. To this aim, primary liver cells (40 Tumor and 40 Non-Tumor cells) obtained from resected hepatocellular carcinoma (HCC) tumor tissue and the adjacent non-tumor area (negative control) were analyzed by Raman micro-spectroscopy. Preliminarily, the cells were analyzed morphologically and spectrally. Then, three machine learning approaches, including multivariate models and neural networks, were simultaneously investigated and successfully used to analyze the cells’ Raman data. The results clearly demonstrate the effectiveness of artificial intelligence (AI)-assisted Raman spectroscopy for Tumor cell classification and prediction with an accuracy of nearly 90% of correct predictions on a single spectrum.
Full article
►▼
Show Figures
Open AccessArticle
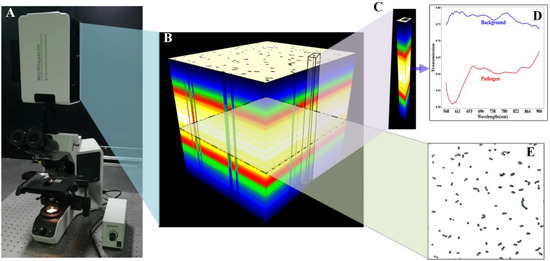
Rapid Identification of Infectious Pathogens at the Single-Cell Level via Combining Hyperspectral Microscopic Images and Deep Learning
by
Chenglong Tao, Jian Du, Junjie Wang, Bingliang Hu and Zhoufeng Zhang
Cited by 13 | Viewed by 3835
Abstract
Identifying infectious pathogens quickly and accurately is significant for patients and doctors. Identifying single bacterial strains is significant in eliminating culture and speeding up diagnosis. We present an advanced optical method for the rapid detection of infectious (including common and uncommon) pathogens by
[...] Read more.
Identifying infectious pathogens quickly and accurately is significant for patients and doctors. Identifying single bacterial strains is significant in eliminating culture and speeding up diagnosis. We present an advanced optical method for the rapid detection of infectious (including common and uncommon) pathogens by combining hyperspectral microscopic imaging and deep learning. To acquire more information regarding the pathogens, we developed a hyperspectral microscopic imaging system with a wide wavelength range and fine spectral resolution. Furthermore, an end-to-end deep learning network based on feature fusion, called BI-Net, was designed to extract the species-dependent features encoded in cell-level hyperspectral images as the fingerprints for species differentiation. After being trained based on a large-scale dataset that we built to identify common pathogens, BI-Net was used to classify uncommon pathogens via transfer learning. An extensive analysis demonstrated that BI-Net was able to learn species-dependent characteristics, with the classification accuracy and Kappa coefficients being 92% and 0.92, respectively, for both common and uncommon species. Our method outperformed state-of-the-art methods by a large margin and its excellent performance demonstrates its excellent potential in clinical practice.
Full article
►▼
Show Figures
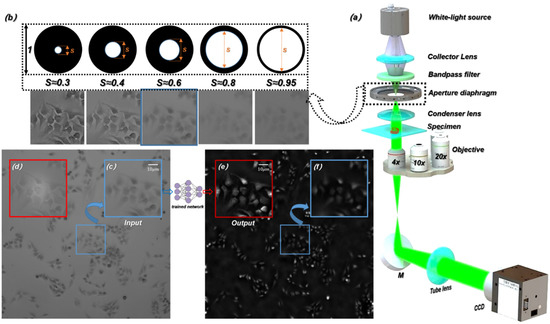
Open AccessReview
Computational Portable Microscopes for Point-of-Care-Test and Tele-Diagnosis
by
Yinxu Bian, Tao Xing, Kerong Jiao, Qingqing Kong, Jiaxiong Wang, Xiaofei Yang, Shenmin Yang, Yannan Jiang, Renbing Shen, Hua Shen and Cuifang Kuang
Cited by 6 | Viewed by 5092
Abstract
In bio-medical mobile workstations, e.g., the prevention of epidemic viruses/bacteria, outdoor field medical treatment and bio-chemical pollution monitoring, the conventional bench-top microscopic imaging equipment is limited. The comprehensive multi-mode (bright/dark field imaging, fluorescence excitation imaging, polarized light imaging, and differential interference microscopy imaging,
[...] Read more.
In bio-medical mobile workstations, e.g., the prevention of epidemic viruses/bacteria, outdoor field medical treatment and bio-chemical pollution monitoring, the conventional bench-top microscopic imaging equipment is limited. The comprehensive multi-mode (bright/dark field imaging, fluorescence excitation imaging, polarized light imaging, and differential interference microscopy imaging, etc.) biomedical microscopy imaging systems are generally large in size and expensive. They also require professional operation, which means high labor-cost, money-cost and time-cost. These characteristics prevent them from being applied in bio-medical mobile workstations. The bio-medical mobile workstations need microscopy systems which are inexpensive and able to handle fast, timely and large-scale deployment. The development of lightweight, low-cost and portable microscopic imaging devices can meet these demands. Presently, for the increasing needs of point-of-care-test and tele-diagnosis, high-performance computational portable microscopes are widely developed. Bluetooth modules, WLAN modules and 3G/4G/5G modules generally feature very small sizes and low prices. And industrial imaging lens, microscopy objective lens, and CMOS/CCD photoelectric image sensors are also available in small sizes and at low prices. Here we review and discuss these typical computational, portable and low-cost microscopes by refined specifications and schematics, from the aspect of optics, electronic, algorithms principle and typical bio-medical applications.
Full article
►▼
Show Figures
Open AccessArticle
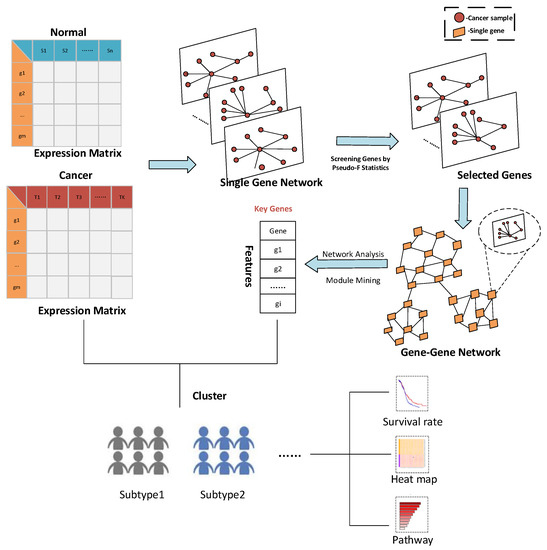
HSSG: Identification of Cancer Subtypes Based on Heterogeneity Score of A Single Gene
by
Shanchen Pang, Wenhao Wu, Yuanyuan Zhang, Shudong Wang, Muyuan Niu, Kuijie Zhang and Wenjing Yin
Cited by 2 | Viewed by 2601
Abstract
Cancer is a highly heterogeneous disease, which leads to the fact that even the same cancer can be further classified into different subtypes according to its pathology. With the multi-omics data widely used in cancer subtypes identification, effective feature selection is essential for
[...] Read more.
Cancer is a highly heterogeneous disease, which leads to the fact that even the same cancer can be further classified into different subtypes according to its pathology. With the multi-omics data widely used in cancer subtypes identification, effective feature selection is essential for accurately identifying cancer subtypes. However, the feature selection in the existing cancer subtypes identification methods has the problem that the most helpful features cannot be selected from a biomolecular perspective, and the relationship between the selected features cannot be reflected. To solve this problem, we propose a method for feature selection to identify cancer subtypes based on the heterogeneity score of a single gene: HSSG. In the proposed method, the sample-similarity network of a single gene is constructed, and pseudo-F statistics calculates the heterogeneity score for cancer subtypes identification of each gene. Finally, we construct gene-gene networks using genes with higher heterogeneity scores and mine essential genes from the networks. From the seven TCGA data sets for three experiments, including cancer subtypes identification in single-omics data, the performance in feature selection of multi-omics data, and the effectiveness and stability of the selected features, HSSG achieves good performance in all. This indicates that HSSG can effectively select features for subtypes identification.
Full article
►▼
Show Figures
Open AccessArticle
ContransGAN: Convolutional Neural Network Coupling Global Swin-Transformer Network for High-Resolution Quantitative Phase Imaging with Unpaired Data
by
Hao Ding, Fajing Li, Xiang Chen, Jun Ma, Shouping Nie, Ran Ye and Caojin Yuan
Cited by 13 | Viewed by 3172
Abstract
Optical quantitative phase imaging (QPI) is a frequently used technique to recover biological cells with high contrast in biology and life science for cell detection and analysis. However, the quantitative phase information is difficult to directly obtain with traditional optical microscopy. In addition,
[...] Read more.
Optical quantitative phase imaging (QPI) is a frequently used technique to recover biological cells with high contrast in biology and life science for cell detection and analysis. However, the quantitative phase information is difficult to directly obtain with traditional optical microscopy. In addition, there are trade-offs between the parameters of traditional optical microscopes. Generally, a higher resolution results in a smaller field of view (FOV) and narrower depth of field (DOF). To overcome these drawbacks, we report a novel semi-supervised deep learning-based hybrid network framework, termed ContransGAN, which can be used in traditional optical microscopes with different magnifications to obtain high-quality quantitative phase images. This network framework uses a combination of convolutional operation and multiheaded self-attention mechanism to improve feature extraction, and only needs a few unpaired microscopic images to train. The ContransGAN retains the ability of the convolutional neural network (CNN) to extract local features and borrows the ability of the Swin-Transformer network to extract global features. The trained network can output the quantitative phase images, which are similar to those restored by the transport of intensity equation (TIE) under high-power microscopes, according to the amplitude images obtained by low-power microscopes. Biological and abiotic specimens were tested. The experiments show that the proposed deep learning algorithm is suitable for microscopic images with different resolutions and FOVs. Accurate and quick reconstruction of the corresponding high-resolution (HR) phase images from low-resolution (LR) bright-field microscopic intensity images was realized, which were obtained under traditional optical microscopes with different magnifications.
Full article
►▼
Show Figures
Open AccessArticle
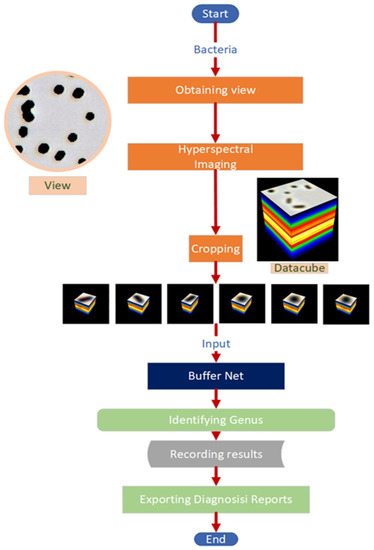
A Deep-Learning Based System for Rapid Genus Identification of Pathogens under Hyperspectral Microscopic Images
by
Chenglong Tao, Jian Du, Yingxin Tang, Junjie Wang, Ke Dong, Ming Yang, Bingliang Hu and Zhoufeng Zhang
Cited by 20 | Viewed by 4216
Abstract
Infectious diseases have always been a major threat to the survival of humanity. Additionally, they bring an enormous economic burden to society. The conventional methods for bacteria identification are expensive, time-consuming and laborious. Therefore, it is of great importance to automatically rapidly identify
[...] Read more.
Infectious diseases have always been a major threat to the survival of humanity. Additionally, they bring an enormous economic burden to society. The conventional methods for bacteria identification are expensive, time-consuming and laborious. Therefore, it is of great importance to automatically rapidly identify pathogenic bacteria in a short time. Here, we constructed an AI-assisted system for automating rapid bacteria genus identification, combining the hyperspectral microscopic technology and a deep-learning-based algorithm Buffer Net. After being trained and validated in the self-built dataset, which consists of 11 genera with over 130,000 hyperspectral images, the accuracy of the algorithm could achieve 94.9%, which outperformed 1D-CNN, 2D-CNN and 3D-ResNet. The AI-assisted system we developed has great potential in assisting clinicians in identifying pathogenic bacteria at the single-cell level with high accuracy in a cheap, rapid and automatic way. Since the AI-assisted system can identify the pathogenic genus rapidly (about 30 s per hyperspectral microscopic image) at the single-cell level, it can shorten the time or even eliminate the demand for cultivating. Additionally, the system is user-friendly for novices.
Full article
►▼
Show Figures
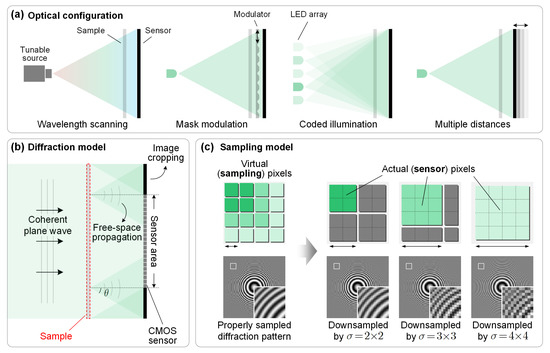
Open AccessFeature PaperArticle
Pixel Super-Resolution Phase Retrieval for Lensless On-Chip Microscopy via Accelerated Wirtinger Flow
by
Yunhui Gao, Feng Yang and Liangcai Cao
Cited by 26 | Viewed by 6563
Abstract
Empowered by pixel super-resolution (PSR) and phase retrieval techniques, lensless on-chip microscopy opens up new possibilities for high-throughput biomedical imaging. However, the current PSR phase retrieval approaches are time consuming in terms of both the measurement and reconstruction procedures. In this work, we
[...] Read more.
Empowered by pixel super-resolution (PSR) and phase retrieval techniques, lensless on-chip microscopy opens up new possibilities for high-throughput biomedical imaging. However, the current PSR phase retrieval approaches are time consuming in terms of both the measurement and reconstruction procedures. In this work, we present a novel computational framework for PSR phase retrieval to address these concerns. Specifically, a sparsity-promoting regularizer is introduced to enhance the well posedness of the nonconvex problem under limited measurements, and Nesterov’s momentum is used to accelerate the iterations. The resulting algorithm, termed accelerated Wirtinger flow (AWF), achieves at least an order of magnitude faster rate of convergence and allows a twofold reduction in the measurement number while maintaining competitive reconstruction quality. Furthermore, we provide general guidance for step size selection based on theoretical analyses, facilitating simple implementation without the need for complicated parameter tuning. The proposed AWF algorithm is compatible with most of the existing lensless on-chip microscopes and could help achieve label-free rapid whole slide imaging of dynamic biological activities at subpixel resolution.
Full article
►▼
Show Figures
Open AccessFeature PaperArticle
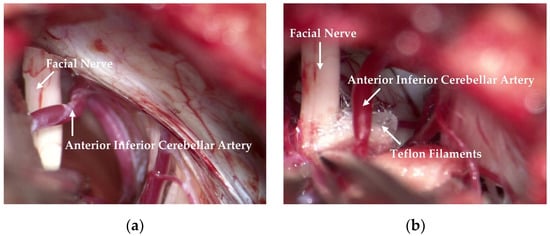
Deep Learning Based Real-Time Semantic Segmentation of Cerebral Vessels and Cranial Nerves in Microvascular Decompression Scenes
by
Ruifeng Bai, Xinrui Liu, Shan Jiang and Haijiang Sun
Cited by 9 | Viewed by 3633
Abstract
Automatic extraction of cerebral vessels and cranial nerves has important clinical value in the treatment of trigeminal neuralgia (TGN) and hemifacial spasm (HFS). However, because of the great similarity between different cerebral vessels and between different cranial nerves, it is challenging to segment
[...] Read more.
Automatic extraction of cerebral vessels and cranial nerves has important clinical value in the treatment of trigeminal neuralgia (TGN) and hemifacial spasm (HFS). However, because of the great similarity between different cerebral vessels and between different cranial nerves, it is challenging to segment cerebral vessels and cranial nerves in real time on the basis of true-color microvascular decompression (MVD) images. In this paper, we propose a lightweight, fast semantic segmentation Microvascular Decompression Network (MVDNet) for MVD scenarios which achieves a good trade-off between segmentation accuracy and speed. Specifically, we designed a Light Asymmetric Bottleneck (LAB) module in the encoder to encode context features. A Feature Fusion Module (FFM) was introduced into the decoder to effectively combine high-level semantic features and underlying spatial details. The proposed network has no pretrained model, fewer parameters, and a fast inference speed. Specifically, MVDNet achieved 76.59% mIoU on the MVD test set, has 0.72 M parameters, and has a 137 FPS speed using a single GTX 2080Ti card.
Full article
►▼
Show Figures
Open AccessFeature PaperArticle
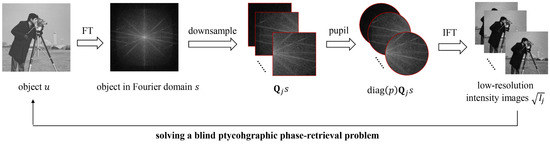
Fourier Ptychographic Microscopy via Alternating Direction Method of Multipliers
by
Aiye Wang, Zhuoqun Zhang, Siqi Wang, An Pan, Caiwen Ma and Baoli Yao
Cited by 35 | Viewed by 4834
Abstract
Fourier ptychographic microscopy (FPM) has risen as a promising computational imaging technique that breaks the trade-off between high resolution and large field of view (FOV). Its reconstruction is normally formulated as a blind phase retrieval problem, where both the object and probe have
[...] Read more.
Fourier ptychographic microscopy (FPM) has risen as a promising computational imaging technique that breaks the trade-off between high resolution and large field of view (FOV). Its reconstruction is normally formulated as a blind phase retrieval problem, where both the object and probe have to be recovered from phaseless measured data. However, the stability and reconstruction quality may dramatically deteriorate in the presence of noise interference. Herein, we utilized the concept of alternating direction method of multipliers (ADMM) to solve this problem (termed ADMM-FPM) by breaking it into multiple subproblems, each of which may be easier to deal with. We compared its performance against existing algorithms in both simulated and practical FPM platform. It is found that ADMM-FPM method belongs to a global optimization algorithm with a high degree of parallelism and thus results in a more stable and robust phase recovery under noisy conditions. We anticipate that ADMM will rekindle interest in FPM as more modifications and innovations are implemented in the future.
Full article
►▼
Show Figures
Open AccessFeature PaperArticle
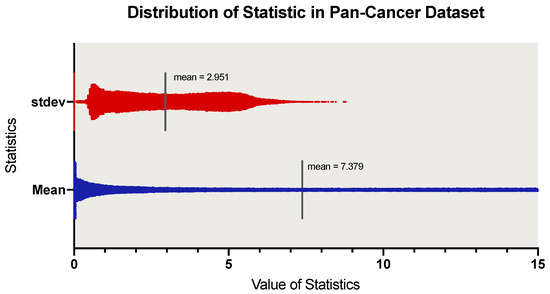
A Novel Attention-Mechanism Based Cox Survival Model by Exploiting Pan-Cancer Empirical Genomic Information
by
Xiangyu Meng, Xun Wang, Xudong Zhang, Chaogang Zhang, Zhiyuan Zhang, Kuijie Zhang and Shudong Wang
Cited by 14 | Viewed by 3695
Abstract
Cancer prognosis is an essential goal for early diagnosis, biomarker selection, and medical therapy. In the past decade, deep learning has successfully solved a variety of biomedical problems. However, due to the high dimensional limitation of human cancer transcriptome data and the small
[...] Read more.
Cancer prognosis is an essential goal for early diagnosis, biomarker selection, and medical therapy. In the past decade, deep learning has successfully solved a variety of biomedical problems. However, due to the high dimensional limitation of human cancer transcriptome data and the small number of training samples, there is still no mature deep learning-based survival analysis model that can completely solve problems in the training process like overfitting and accurate prognosis. Given these problems, we introduced a novel framework called SAVAE-Cox for survival analysis of high-dimensional transcriptome data. This model adopts a novel attention mechanism and takes full advantage of the adversarial transfer learning strategy. We trained the model on 16 types of TCGA cancer RNA-seq data sets. Experiments show that our module outperformed state-of-the-art survival analysis models such as the Cox proportional hazard model (Cox-ph), Cox-lasso, Cox-ridge, Cox-nnet, and VAECox on the concordance index. In addition, we carry out some feature analysis experiments. Based on the experimental results, we concluded that our model is helpful for revealing cancer-related genes and biological functions.
Full article
►▼
Show Figures